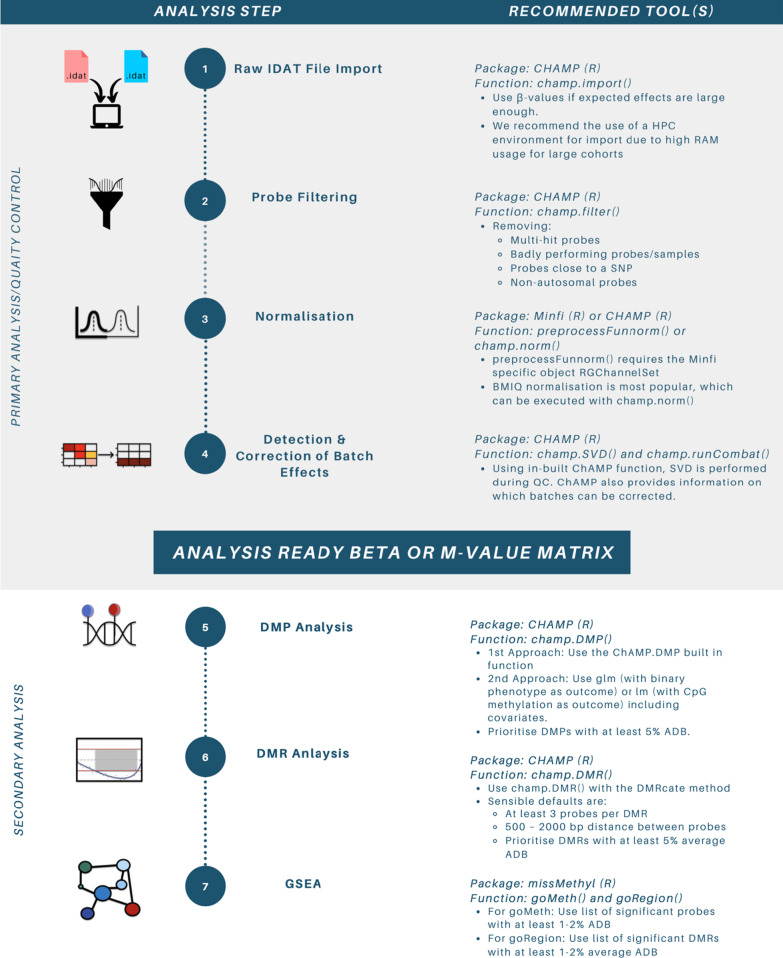
Fig. 4.
Steps and tools for primary EWAS analysis steps. Listed tools include ChAMP (https://bioconductor.org/packages/release/bioc/html/ChAMP.html), Minfi (https://bioconductor.org/packages/release/bioc/html/minfi.html) and missMethyl (http://bioconductor.org/packages/release/bioc/html/missMethyl.html). Abbreviations: EWAS = epigenome-wide association study, ChAMP = chip analysis methylation pipeline, HPC = high performance computer, RAM = random access memory, SNP = single nucleotide polymorphism, BMIQ = beta mixture quantile, SVD = singular value decomposition, QC = quality control, DMR = differentially methylated region, ADB = absolute deta beta