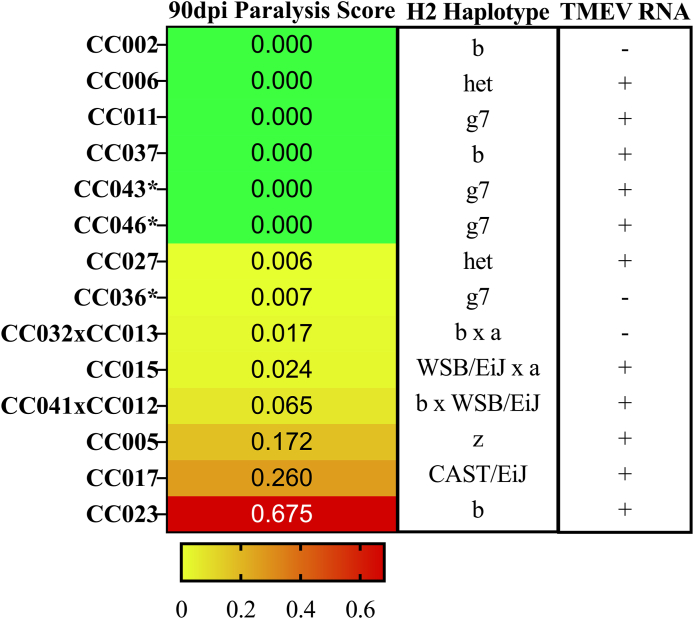
Fig. 4.
CC strains and their respective H2 haplotypes and TMEVpresence/absence are categorized according to observed paralysis scores. CC strains are arranged by increasing paralysis frequency scores (green to red gradient). We identified the H2 haplotypes for each strain based on the founder strains from which the H2 complex was inherited, to demonstrate that H2 haplotype alone does not influence phenotypic severity. TMEV RNA presence was measured via relative levels of the TMEV polyprotein sequence AAA47930 within sham and infected mice. Those strains with detectable TMEV RNA within the CNS tissue are denoted with “+”; “-” denotes those strains with no detectable TMEV RNA. A complete RNA sequencing report of relative TMEV RNA levels are found in B1. ∗strains not included in (Eldridge et al., 2020). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)