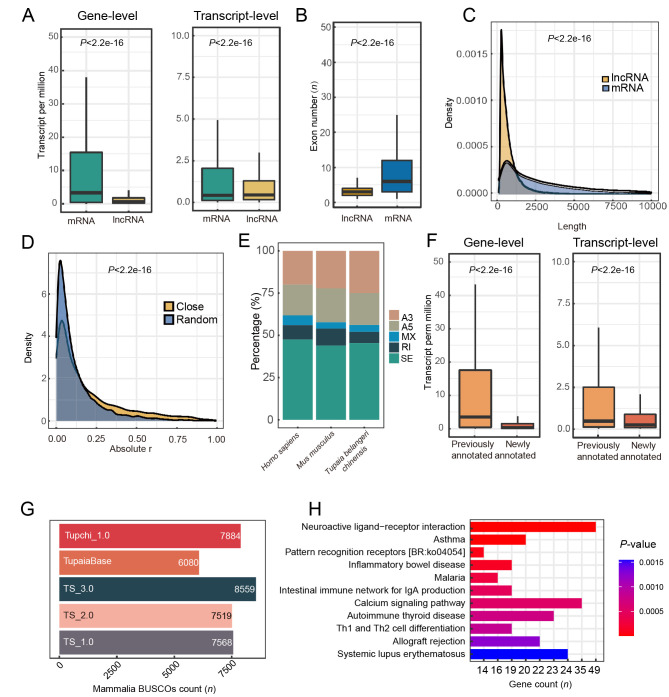
Figure 2.
Characteristics of tree shrew TS_3.0 transcripts
A: Expression level of mRNAs is greater than that of lncRNAs at gene and transcript levels. B: mRNA transcripts have a higher number of exons than lncRNA transcripts. C: Average length of mRNA transcripts is longer than that of lncRNA transcripts. Density plot was drawn based on kernel density and statistical analysis was performed with Wilcoxon rank-sum test. D: Tree shrew lncRNAs exert cis-regulatory function on expression of proximal mRNAs. Close, mRNA and lncRNA pairs neighboring each other on pseudochromosome of tree shrew. Random, randomly selected mRNA and lncRNA pairs distant from each other in the genome. E: Percentages of alternative splicing events in reference genome annotations of tree shrew (TS_3.0), mouse (GRCm39), and human (GRCh38.p13). SE, skipped exon; A5, alternative 5’ splice site; A3, alternative 3’ splice site; MXE, mutually exclusive exons; RI, retained intron. F: Expression level of newly annotated coding genes is lower than that of previously annotated genes at gene and transcript levels. G: BUSCO evaluation of different tree shrew genome annotations showing that current version (TS_3.0) is superior. H: Pathway enrichment analysis of newly annotated genes showing enrichment in 11 pathways (Padjust<0.05). Values in A, B and F are presented as a boxplot, and statistical analyses were performed by Wilcoxon rank-sum test.