FIGURE 1.
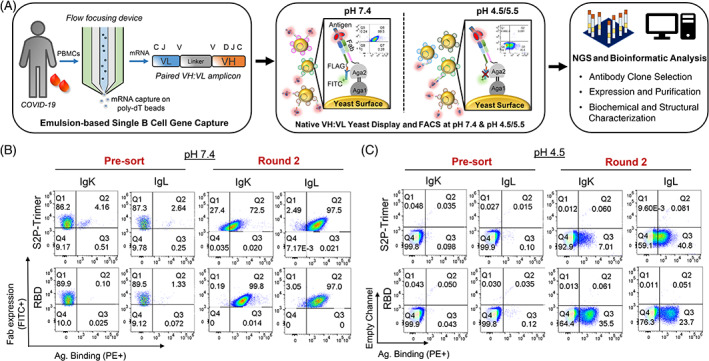
Functional analysis and screening of natively paired variable heavy:variable light (VH:VL) antibody repertoires from a convalescent COVID‐19 patient. (A) Natively paired VH:VL antibody repertoires were generated from a COVID‐19 convalescent donor for functional screening via yeast display. Left: Single B‐cell capture using a flow‐focusing device generating microdroplets consisting of a single B cell, lysis buffer, and oligo‐dT magnetic beads. Single cells are emulsified and mRNA is captured onto magnetic beads used as template for overlap extension RT‐PCR reactions to produce linked antibody VH and VL cDNAs; Center: Natively paired VH:VL cDNA amplicon libraries are cloned into a yeast display vector and expressed as Fabs for flow cytometric cell sorting (FACS) analysis at pH 7.4 and pH 4.5/5.5. Fab expression at pH 7.4 is detected by an anti‐FLAG FITC‐conjugated mAb; antigen binding is detected via PE‐conjugated antigen probes. Fab expression is not measured at pH 4.5/5.5 due to markedly reduced binding affinity between FLAG expression tag. Screening of yeast‐displayed natively paired VH:VL libraries against the SARS‐CoV‐2 spike glycoprotein with two stabilizing proline mutations in S2 domain (S2P‐Trimer) and RBD antigens is shown via FACS at (B) pH 7.4 and (C) pH 4.5. IgK stands for antibodies encoding kappa light chains, and IgL for antibodies with lambda light chains. Substantial enrichment in antigen binding was observed after three rounds of sorting both at pH 7.4 and 4.5 compared to the presort/input libraries. See Figure S1 for sorting and enrichment at pH 5.5, and for an evaluation of expression and binding at pH 7.4 of yeast libraries sorted at pH 4.5/5.5
