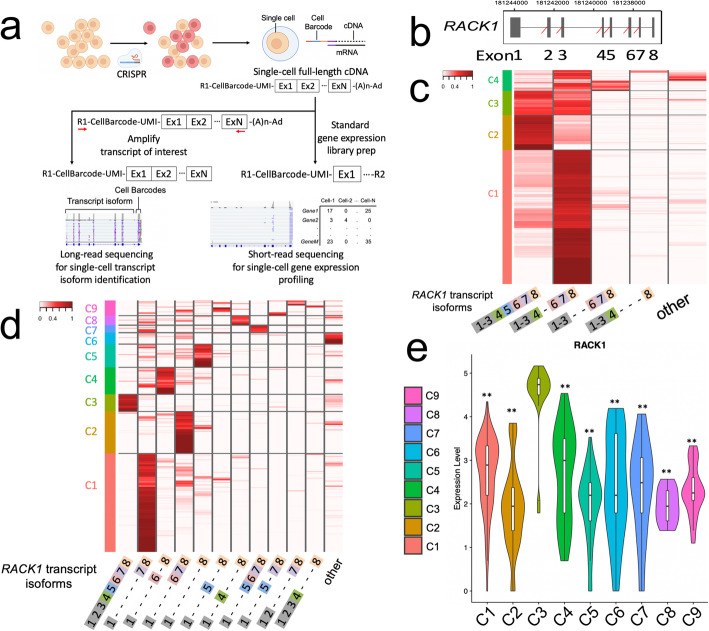
Fig. 1.
a Overview of single-cell short/long-read integration strategy of RACK1 targeting. b Structure of RACK1 transcript. c, d Two heatmaps showing the proportion of each transcript isoform (x-axis) with each cell (y-axis) for either the single (c) or multiplexed (d) CRISPR edits respectively. For panel c, hierarchical clustering was conducted on the cells subject to a single CRISPR edit of exon 5. Cluster 2 has the intact RACK1 isoform with all exons. Cluster 1 cells demonstrate exon 5 skipping. For panel d, hierarchical clustering was conducted on cells subject to multiplexed edits across different exons. Cluster 3 indicates the cells with an intact RACK1 transcript. The remaining clusters demonstrate cells with exon skipping. e Expression level of RACK1 in each cell cluster denoted in panel d. P values were calculated for each cluster in comparison with Cluster 3 (C1 P = 5.7e−12, C2 2.6e−11, C4 9.9e−11, C5 1.6e−10, C6 1.1e−07, C7 9.5e−06, C8 2.5e−07, C9 1.1e−08 ; two-sided t-test)