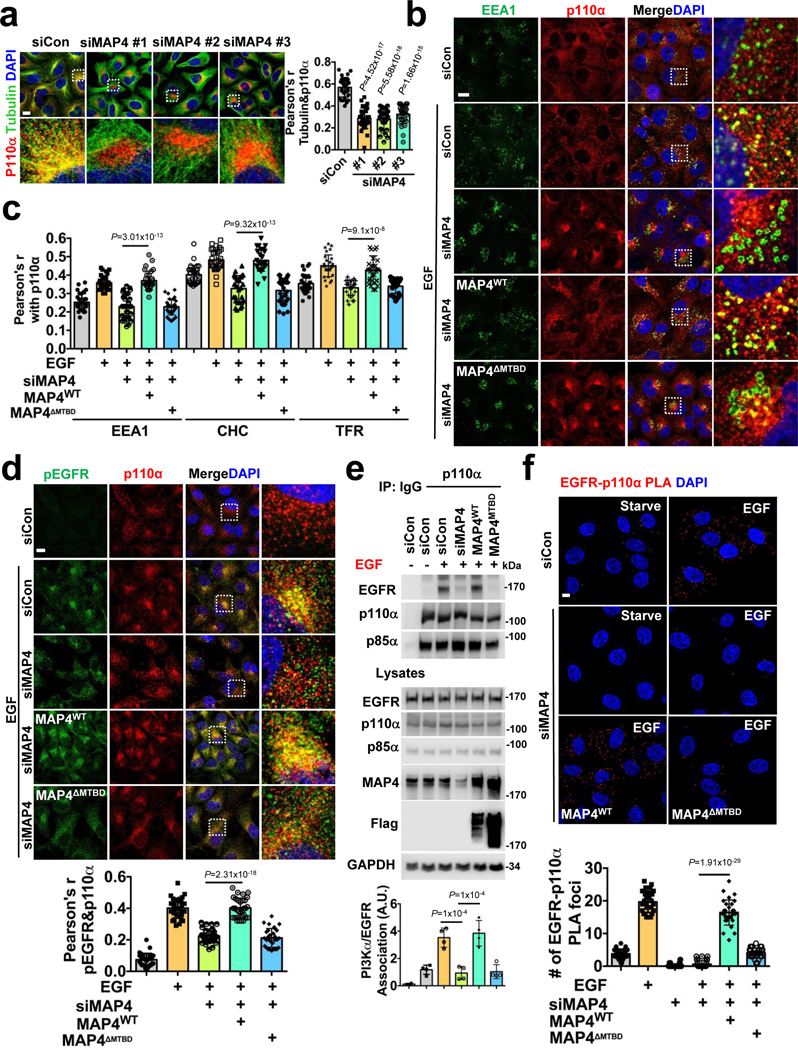
FIGURE 5: MAP4 is Required for PI3Kα Vesicle Distribution along Microtubules and its Association with Activated Receptors.
a, MAP4 loss affects PI3Kα vesicle distribution along microtubules. Three different individual siRNAs were used for MAP4 knockdown in MDA-MB-231 cells followed by immunofluorescence staining. The co-localization of p110α vesicles and tubulin were quantified by Pearson’s r. Scale bar, 5 μm; Error bars denote mean±SD; n=30 cells from representative experiments
b, c, MAP4 is required for the endosomal localization of PI3Kα vesicles. The siRNAs targeting the 3’ UTR region of MAP4 (siMAP4#3) were used to knockdown endogenous MAP4 in MDA-MB-231 cells expressing WT or MTBD deletion mutant of MAP4. 48–72 hrs post-transfection, cells were stimulated with EGF and processed for immunofluorescence study using antibodies specific for endogenous p110α and endosomal markers (EEA1). Co-localization with CHC and TFR is shown in Figure S7a-b. The co-localization of p110α and EEA1/CHC/TFR was quantified by Pearson’s r. Scale bar, 5 μm; Error bars denote mean±SD, n=30 cells from representative experiments
d, e, f, MAP4 knockdown impairs the association of PI3Kα with activated EGFR. siRNAs targeting the 3’ UTR region of MAP4 were used to knockdown endogenous MAP4 in MDA-MB-231 cells expressing WT or the MTBD deletion mutant of MAP4. 48–72 hrs post-transfection, cells were stimulated with EGF and processed for immunofluorescence study using antibodies specific for endogenous p110α and activated EGFR. The co-localization between p110α and pEGFR was quantified by Pearson’s r. Similarly, the effect of MAP4 knockdown in the association of PI3Kα with EGFR was analyzed by co-immunoprecipitation assay and proximity ligation assay. Scale bar, 5 μm; Error bars denote mean±SD; n=30 cells from representative experiments (d and f), n=4 independent experiments (e)
Unprocessed_Western_Blots_Fig5; Statistical_Source_Data_Fig5