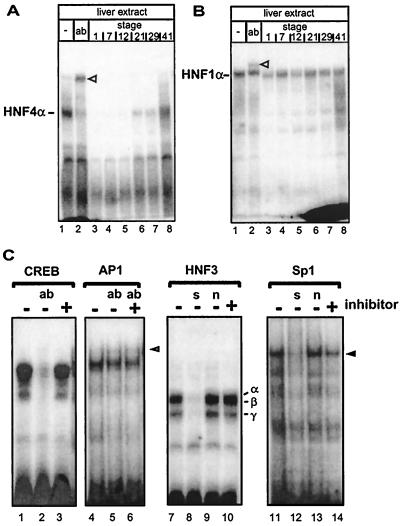
FIG. 1.
Identification of an HNF4 inhibitor in Xenopus eggs that gradually disappears during development. (A) Xenopus liver extract (3 μl) was incubated with the labeled oligonucleotide containing the HNF4 binding site of the human apolipoprotein B promoter and analyzed in a gel retardation assay. Monoclonal antibody H4/55, specific for HNF4α (32), is in lane 2 (ab). Lanes 3 to 8 contain in addition 1 μl of embryonic extracts from the developmental stages given (27). The HNF4α-DNA complex is indicated, and the supershifted complex containing the antibody is marked with an open triangle. (B) Gel retardation assay essentially as in panel A, but using the HNF1 binding site of the albumin promoter in combination with monoclonal antibody XAD5, specific for HNF1α (1). (C) Gel retardation assay using rat liver nuclear extract and a CREB binding site (lanes 1 to 3), an AP1 site (lanes 4 to 6), an HNF3 site (lanes 7 to 10), and an Sp1 site (lanes 11 to 14). Protein-free HNF4 inhibitor preparation (1 μl) was added to the reactions as indicated. We used this protein-free inhibitor preparation to avoid a change in protein concentration that may alter the DNA binding in the gel retardation assays. To identify protein-DNA complexes, antibodies (ab) were added as indicated: lane 2, 2 μg of antibody CREB-1 (Santa Cruz; C-21) directed against the DNA-binding domain of CREB; lanes 5 to 6, 2 μg of antibody against c-Fos (Santa Cruz; K-25). The complex supershifted by antibody to c-Fos (K-25) is marked by an open triangle in lane 6. In lanes 8 and 12, a 100-fold excess of unlabeled specific competitor (s) was added (HNF3 site and Sp1 site, respectively). In lanes 9 and 13, a 100-fold excess of unlabeled nonspecific competitor (n) was included in the reaction (Sp1 and HNF3 site, respectively). HNF3α, -β, and -γ protein-DNA complexes are indicated in lane 10 (18), and the Sp1 protein-DNA complex is marked by an arrowhead in lane 14 (22).