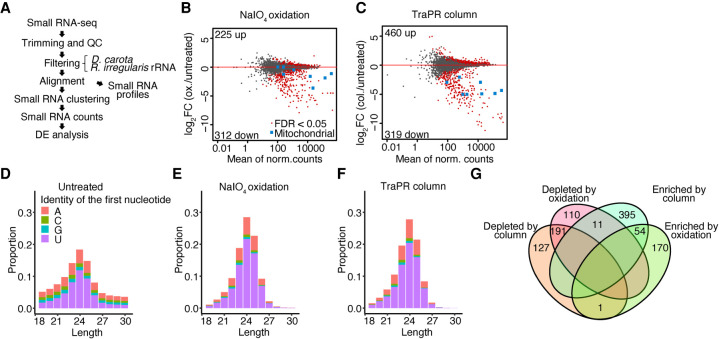
Figure 5.
Isolation of 2′-O-methylated and Argonaute-loaded small RNA. (A) Schematic representation of the sRNA-seq analysis pipeline used in this study. (B,C) Plot of mean normalized counts against log2 fold change (log2FC) in sRNAs sequenced following enrichment using NaIO4 oxidation (B) or TraPR column purification (C), compared to sRNAs sequenced following no treatment. Points represent individual sRNA loci. Significantly differentially expressed loci are shown in red. Mitochondria-derived sRNA loci are shown in blue. (D–F) Length distribution and first nucleotide bias of the sRNAs in untreated (D), NaIO4-treated (E), and TraPR column-extracted (F) sRNA libraries. (G) Significantly differentially expressed sRNA loci. sRNAs enriched by NaIO4 or column-purification were induced compared to expression in untreated samples, and sRNAs depleted by NaIO4 or column-purification were down-regulated.