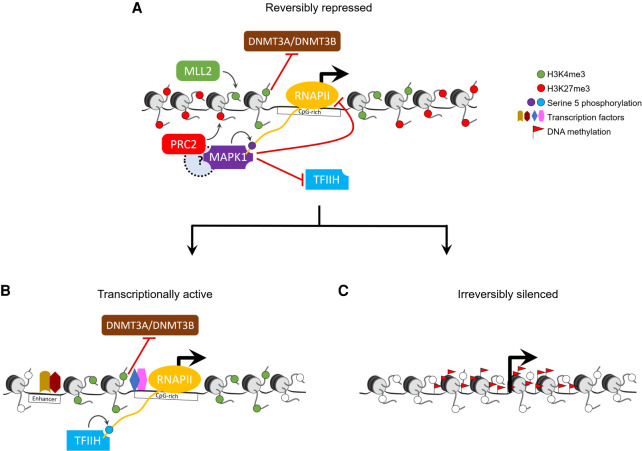
Figure 8.
Model for bivalent chromatin maintaining epigenetic plasticity by protecting gene promoters from irreversible silencing while maintaining a reversibly repressed state. (A) H3K4me3, catalyzed primarily by the MLL2/COMPASS complex, protects CpG-rich bivalent promoters from DNA methylation by repelling de novo methyltransferases DNMT3A and DNMT3B. Bivalent promoters, by virtue of their overlapping CGIs, are more averse to assembling into nucleosomes compared to other genomic DNA (Ramirez-Carrozzi et al. 2009); consequently, bivalent promoters are relatively nucleosome-deficient, intrinsically accessible, and transcriptionally permissive (Deaton and Bird 2011; Mas et al. 2018). The assembly of transcription machinery at the promoter triggers PRC2 to recruit—either directly or indirectly through a yet-to-be-determined mechanism—reinforcement in the form of MAPK1 (ERK2), which, in lieu of the usual TFIIH, phosphorylates serine 5 on a particular (or a set of) RNAPII CTD heptad repeat(s) (Tee et al. 2014). The presence of MAPK1 and/or the ensuing serine 5 phosphorylation is refractory to transcription as it antagonizes the recruitment of the TFIIH complex, which, besides its role in phosphorylating serine 5 on RNAPII, is necessary to unwind promoter DNA to form a transcription bubble for RNA synthesis. (B) Activation of transcription at bivalent genes likely occurs only upon loss of PRC2 activity and thus MAPK1 activity, followed by binding of appropriate transcription factors at promoters and/or enhancers. (C) Loss of H3K4me3 at CpG-rich bivalent promoters make them more susceptible to aberrant DNA methylation during aging and in diseases such as cancer.