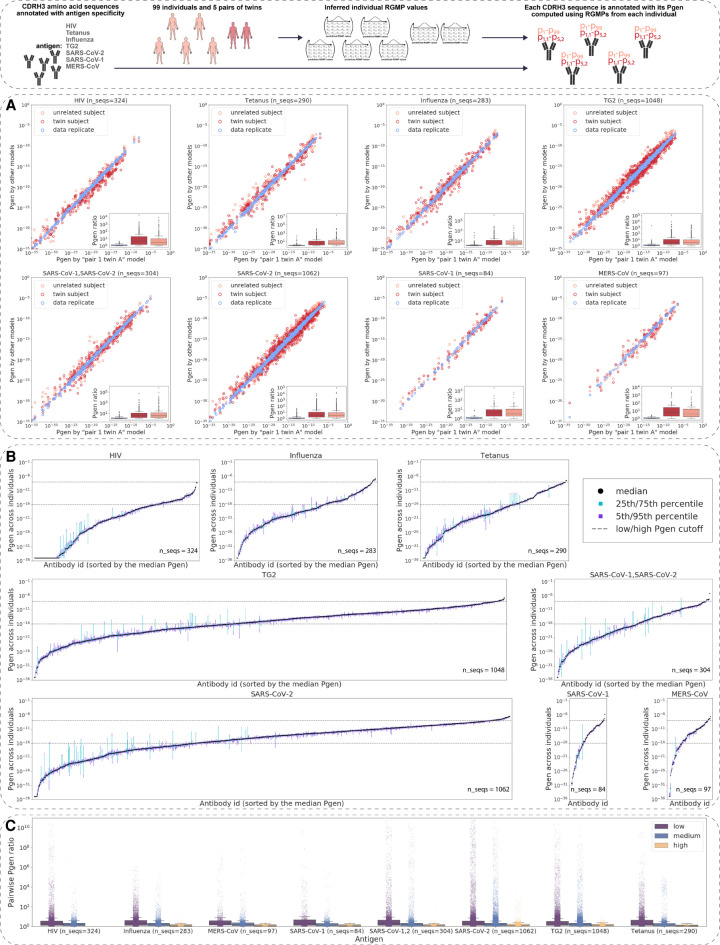
Figure 4.
Generation probabilities of antigen-annotated immunoglobulins (CDRH3 sequences) vary by several orders of magnitude within the human population. (Inset) For a data set of CDRH3 amino acid sequences annotated with antigen specificity, we computed Pgens using a set of RGMs corresponding to N different experimental samples. Each CDRH3 sequence is thus annotated with N Pgens. (A) Pgens of antibody CDRH3 amino acid sequences (annotated with antigen specificity) computed using RGMs corresponding to samples of different levels of immunogenetic similarity: a pair of data replicate models, a pair of models from twin individuals, and a pair of models from unrelated individuals. The x-axis always stands for the Pgen as computed with the model corresponding to the pair 1 twin A individual from the HUMAN2 data set. The y-axis corresponds to the Pgen as computed with the other model in the pair (data replicate or twin/unrelated subject). The boxplots show the distribution of the min(x,y)/max(x,y) ratios, that is, the pairwise difference of Pgens. (B) For each CDRH3 amino acid sequence, we calculated its Pgen as determined by the models corresponding to the 99 individuals from the HUMAN3 data set. The x-axis itemizes each of the CDRH3 sequences tested; the y-axis denotes the fifth, 25th, 50th, 75th, and 95th percentiles of the 99 Pgens of each CDRH3. (C) Pairwise ratios of the Pgens from B by antigen. For each antigen, we divided the CDRH3 amino acid sequences into three groups depending on the sequence's median Pgen across individuals: low (median Pgen < 10−16), medium (10−16 ≤ median Pgen < 10−8), and high (10−8 ≤ median Pgen).