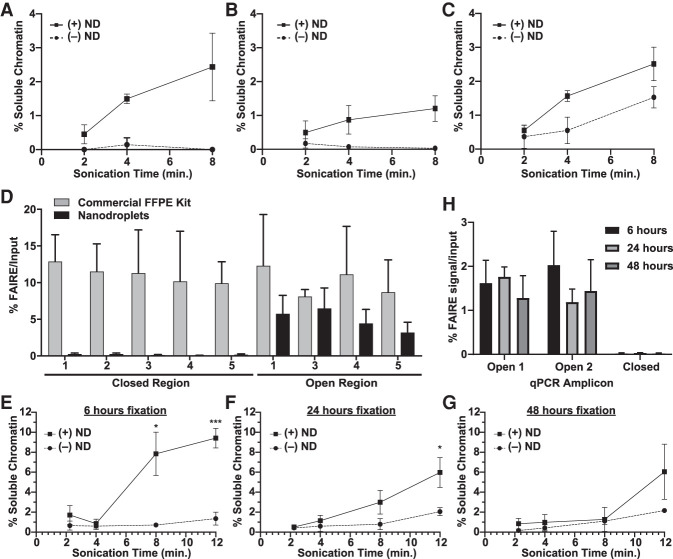
Figure 3.
Nanodroplets facilitate the extraction of chromatin from archival tissue. Percent soluble chromatin from one 10-µm section of UM-RC-2 (A), EWS894 (B), and MDA-MB-231 (C) following sonication with (solid) or without (dashed line) for indicated. (D) FAIRE-qPCR was performed on chromatin prepared from EWS894 xenograft tumor tissue with a commercial FFPE chromatin preparation kit (eight 10-µm sections, gray bars) or with nanodroplets (one 10-µm section, black bars). The y-axis indicates the percent FAIRE signal compared to input signal by qPCR. (Open) regions of known accessible chromatin; (Closed) regions of known inaccessible chromatin. (E–H) Xenograft tumor tissue derived from MCF-7 cells was fixed for 6 h (E), 24 h (F), or 48 h (G), and one, 10-μm section of each was deparaffinized, rehydrated, and sonicated with (solid line) or without (dashed line) nanodroplets for indicated time (x-axis, minutes). Percent soluble chromatin (y-axis) was measured by comparing the quantity of DNA in the soluble fraction to that of the insoluble pellet fraction. (H) FAIRE-qPCR was performed on one, 10-µm tissue section sonicated for 8 min with nanodroplets for each fixation time point. The y-axis indicates the percent FAIRE signal compared to input signal by qPCR. (Open) regions of known accessible chromatin; (Closed) regions of known inaccessible chromatin.