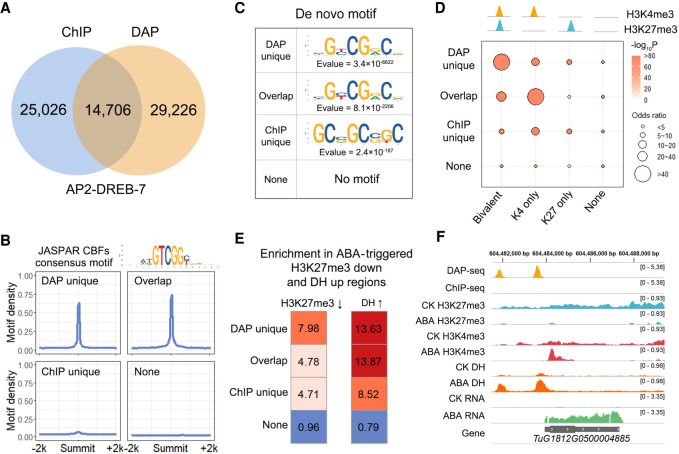
Figure 2.
Concordance of DAP-seq and ChIP-seq peaks. (A) Venn diagrams showing the overlap between ChIP-seq peaks, DAP-seq peaks of AP2-DREB-7, and DHS. (B) Average number of AP2 motifs (bin size = 50 bp) within a 4-kb window centered on common and unique peak summits. The CBF binding motif in the JASPAR plant database is used because CBF is the orthologous gene of Tu AP2-DREB-7 in Arabidopsis. Four CBF binding motifs in the JASPAR database were merged into a consensus motif. (C) Motifs de novo identified from common and unique peaks. (D) Enrichment of histone marks in common and unique peaks. H3K4me3 and H3K27me3 overlapping (bivalent) peaks and unique peaks (K4-only and K27-only) were used for the analysis. (E) Enrichment of DAP-seq unique peaks in H3K27me3 down-regulated and DNase I hypersensitivity (DH) up-regulated regions by ABA. The MAnorm package (Shao et al. 2012) was used for the quantitative comparison of H3K27me3 ChIP-seq and DHS signals between samples. (F) Genomic tracks illustrating the coincidence between DAP-seq unique binding and ABA-induced chromatin accessibility and reduced H3K27me3. TuG1812G0500004885 is a gene with LRR domains which may relate to biotic or abiotic stress responses.