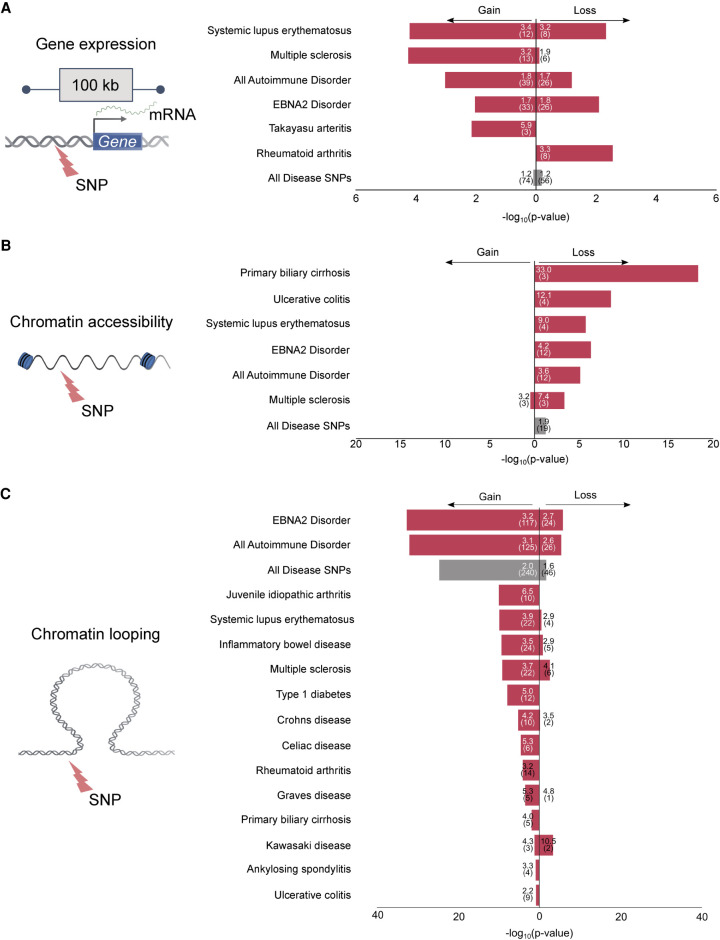
Figure 5.
Intersection of EBNA2-dependent gene regulatory mechanisms and disease-associated genetic variants. The bar plots indicate the significance of the intersection (RELI negative log10 adjusted P-value). For all analyses, 19 autoimmune diseases were individually tested, along with a set of variants from all 19 autoimmune diseases (“All Autoimmune Disorders”), a set containing variants from the nine “EBNA2 Disorders” from our previous study (Harley et al. 2018), and a set containing 176 diseases and phenotypes (“All Disease SNPs”). Only diseases with at least one significant result (three or more overlaps, corrected P-value < 0.05) are shown. Autoimmune diseases are indicated with red bars. Fold enrichment and number of overlaps are indicated inside the bars. (A) Significant intersection between EBNA2-dependent differentially expressed genes and disease-associated variants: (Gain) up-regulated genes; (Loss) down-regulated genes. (B) Significant intersection between EBNA2-dependent chromatin accessibility and disease-associated variants: (Gain) newly opened chromatin; (Loss) newly closed chromatin, relative to uninfected. (C) Significant intersection between EBNA2-dependent chromatin looping and disease-associated variants: (Gain) new looping events; (Loss) loss of looping events.