Figure 1.
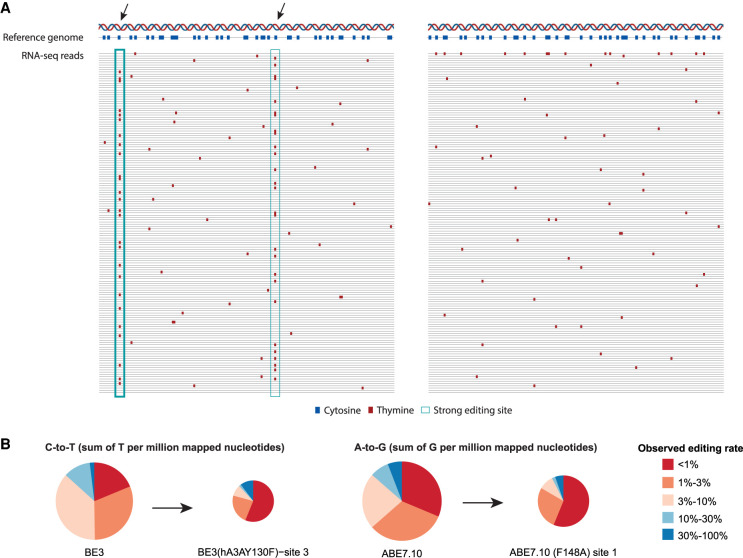
Most RNA mutations due to off-target base editing are nonspecific. (A) Engineered guided deaminases may target efficiently some off-target locations (marked with arrows). These strongly edited sites result in a DNA-RNA mismatch seen in a large fraction of RNA molecules, resembling the mismatch profile observed at genomic polymorphism and clonally selected somatic mutation sites. In parallel, nonspecific off-target base editing activity affects multiple additional sites but a small fraction of RNA molecules per site. However, due to their large number, the total number of nonspecific events could surpass that of specific editing sites, as illustrated in the figure. Hyperedited reads, where multiple mismatches are seen in the same read (right panel, top read), occasionally appear. They provide a strong indication of off-target activity but account for a small minority of mismatches. (B) Relative contribution of editing sites to the editing index, by their observed editing rates, for two engineered base editors. Most off-target activity occurs at weakly edited sites in agreement with the scenario depicted in panel A. Pie area is proportional to the number of detected off-target events. Four base editors from Study H are shown (two cytidine deaminases: BE3, BE3 [hA3AY130F]−site 3; two adenine base editors: ABE7.10 and ABE7.10 [F148A]−site 1). See Supplemental Figure S1 for similar data for all enzymes.