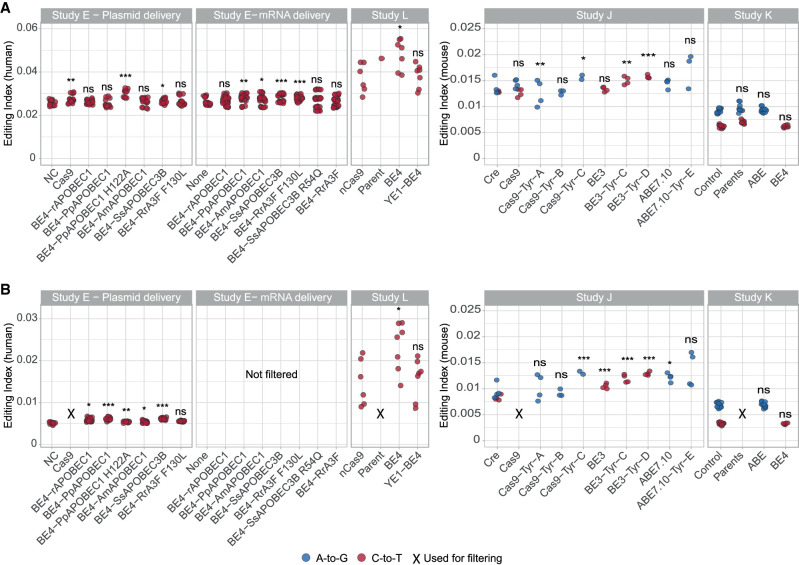
Figure 3.
Off-target DNA editing activity following introduction of base editors. (A) The editing index is a global measure of editing activity, quantifying the fraction (percent) of the DNA nucleotides exhibiting a mismatch with respect to the reference genome. Common polymorphic sites are excluded. Four studies are analyzed (E, L, J, K), examining adenine base editors and cytidine deaminases. For each enzyme, the relevant indices (A-to-G, blue; and C-to-T, red) are calculated over the coding region. The relevant index (A-to-G and C-to-T for adenine and cytidine deaminases, respectively) is compared with control samples (Study E: NC/None; Study L: nCas9; Study J: Cre; Study K: Control). A significant increase is observed for nine enzymes. (B) In order to suppress baseline contributions to the index due to genomic polymorphisms unrelated to the base editor, we repeated the calculation excluding mismatch sites appearing in at least half of the untreated samples not used for the statistical tests (Study E: Cas9; Study L: parent; Study J: Cas9; Study K: parents). Ten of the 15 examined editors exhibit a significant increase in the index, indicating off-target DNA editing. In Study E (mRNA delivery), there were no samples suitable for excluding shared SNVs, and therefore filtering was not applied. Two-sided t-test for log(index); (*) P < 0.05, (**) P < 0.01, (***) P < 0.001. The data per sample are available in Supplemental Table S4, and exact P-values are presented in Supplemental Table S5.