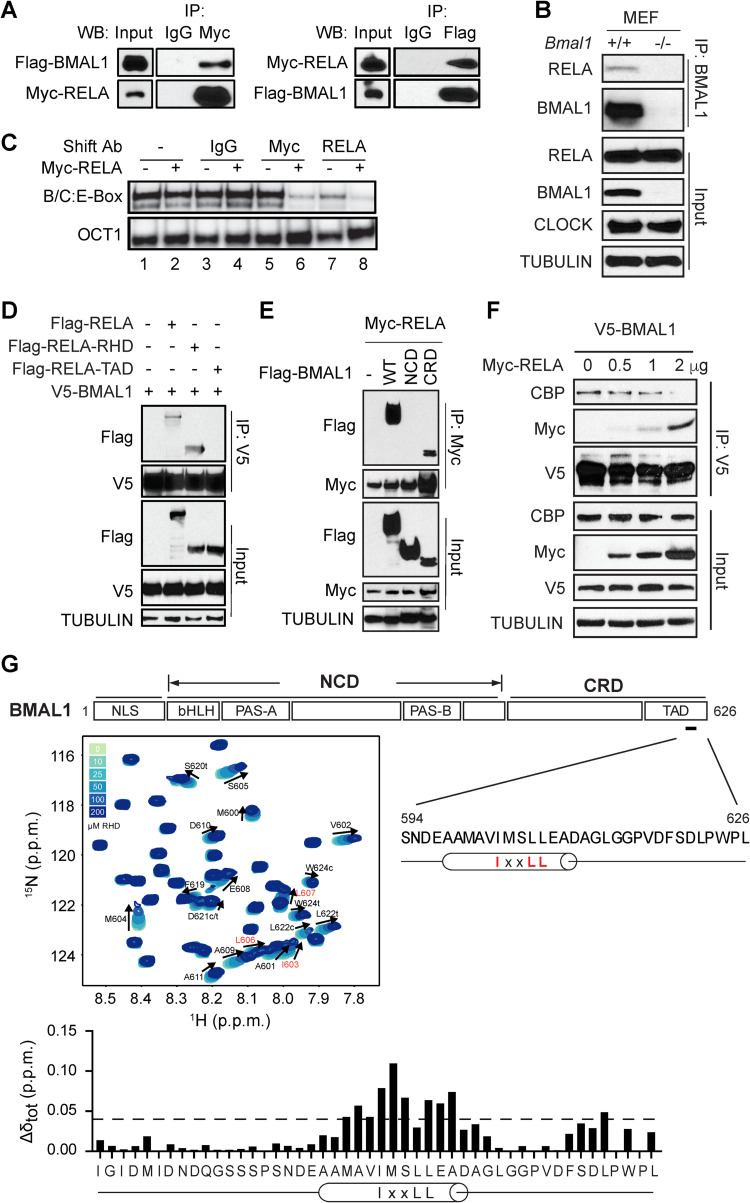
Fig 5. The RELA RHD interacts with the C-terminal transactivation domain of BMAL1.
(A-B) Co-immunoprecipitation assays detect the interaction between RELA and BMAL1 in co-transfected 293T cells (A) and between the endogenous proteins in fibroblasts (B). The intrinsic interaction between RELA and BMAL1 is independent on CLOCK. (C) RELA associates with the BMAL1/CLOCK:E-box complex, indicative of a larger complex formation which caused a supershift. Oct1 oligonucleotide was used as control. The complex was reduced by the anti-Myc antibody, likely caused by a supershift that was not revealed in the gel. (D-E) Co-immunoprecipitation and Western blot analysis to detect the interaction between the RELA RHD and the BMAL1 CRD. 293T cells were co-transfected with tagged BMAL1 and RELA proteins as indicated. (F) Co-immunoprecipitation and Western blot analysis to show competition for interaction with BMAL1 between CBP and RELA. (G) Backbone chemical shift perturbations (Δδtot) of 15N-BMAL1 C-terminal TAD with stoichiometric RELA RHD. Top panel: Schematic diagram of domain structure of BMAL1. The N-terminal core domain (NCD) contains the bHLH, PAS-A and PAS-B domains. The C-terminal regulatory domain (CRD) contains a transactivation domain (TAD) near the C-terminus featuring the highly conserved IxxLL helical motif. Residues within the IxxLL helical motif showed chemical shifts, indicative of interaction with RELA. Middle: Central region of 15N-1H HSQC spectra showing titration of 100 μM 15N-BMAL1 TAD in the presence of increasing concentrations of the RELA RHD (light to dark blue; solid arrow). Peak assignments are indicated for residues in the BMAL1 TAD that undergo chemical shift perturbation upon the addition of RELA RHD. Bottom: Quantification of backbone chemical shift perturbations (ΔδTOT) of 15N-BMAL1 TAD from the titration point with 200 μM RELA RHD. Dashed line, ΔδTOT significance cutoff of 0.04 p.p.m. (parts per million).