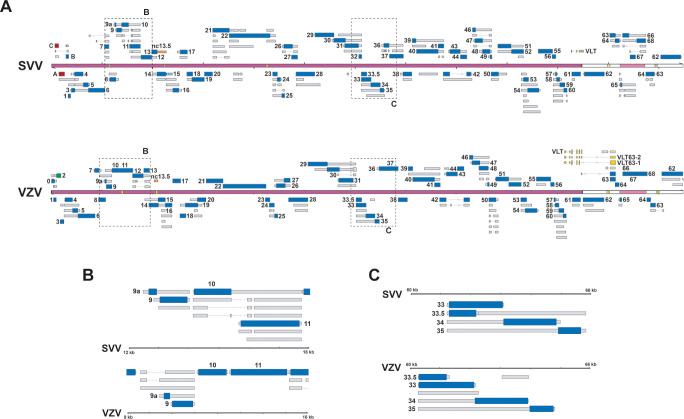
Fig 9. Genome-wide comparison of transcript structures in SVV and VZV.
(A) Alignment of SVV and VZV genomes with their respective annotation of the transcriptome. Canonical ORFs in both viruses are highlighted in blue, with UTRs (thin box, grey) and CDS (wide box), whereas transcript isoforms are given in grey. Unique SVV ORFs are indicated in red, unique VZV ORFs in green, conserved ncRNA 13.5 in orange and VLT in yellow. Double black lines represent the SVV/VZV genome, with the long unique region (UL) in purple fill, short unique region (US) in pink and internal/terminal repeat sequences (IRS/TRS) no fill, and the reiterative repeat regions R1 to R4 and both copies of OriS are indicated as yellow boxes on the genome track. Dashed boxes indicate areas shown as enlargement in (B) and (C). (B) Selected region of RNA9-11 in both viruses where transcript structures differ. (C) Selected region of RNA33-35 in both viruses where transcript structures are similar.