Figure 4.
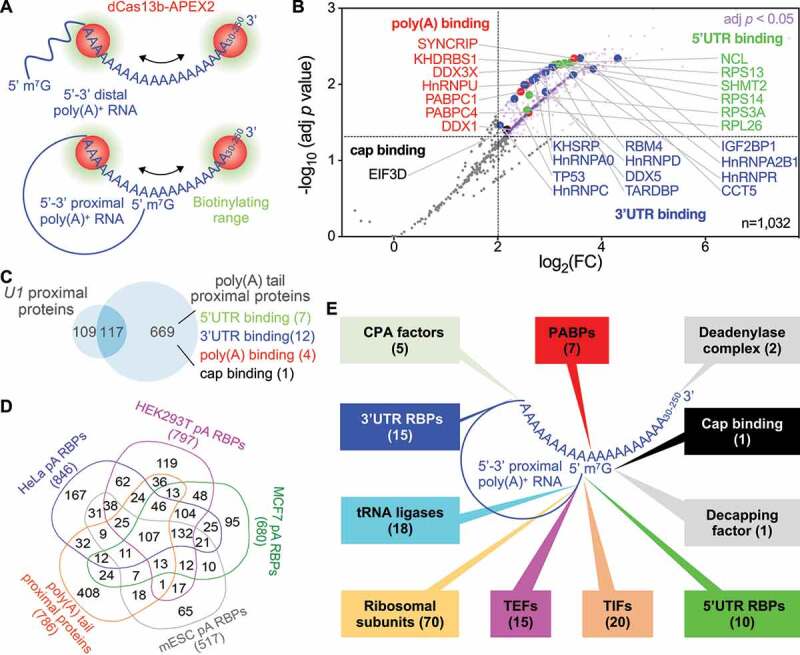
RPL-MS analysis of poly(A) tail proximal proteins in HEK293T cells. (a) Application of RPL to poly(A) tails. In the presence of gRNA, the RPL protein (dCas13b-APEX2) is directed to poly(A) tails ranging from 30 nt up to ~250 nt. In the 5ʹ-3ʹ distal model, RPL will detect poly(A) binding proteins and 3ʹUTR binding proteins that bind proximal to poly(A) tail within the range of biotinylation. In the 5ʹ-3ʹ proximal model, RPL will also identify cap-binding proteins and 5ʹUTR binding proteins that bind proximal to the cap and lie within the biotinylating range. (b) Volcano plot shows RPL-labelled proteins in HEK293T cells. For each protein, the poly(A)/poly(U) iBAQ ratio reflects the enrichment of identified protein in poly(A) gRNA cells compared with poly(U) gRNA transfected cells. RPL-MS identified 786 proteins (light purple dots) as significantly enriched (Benjamini-Hochberg-adjusted p < 0.05 and log2FC > 2). Each data point represents the average value from biological triplicates. Red dots represent poly(A) binding proteins, blue dots 3ʹUTR binding proteins, green dots 5ʹUTR binding proteins, and the black dot denotes a cap binding protein. (c) Venn diagram shows the comparison between poly(A) tail and U1 RPL experiments. KEGG pathways enriched in shared proteins (FDR < 0.01). Authentic categories of proteins associated with poly(A) tail are indicated and number of proteins are shown in brackets. (d) Venn diagram shows the comparison of proteins associated with poly(A)+ RNA in different cells. Numbers below each group represent the size. (e) Summary of expected categories of poly(A) tail proximal proteins that support 5ʹ-3ʹ proximity and the role of poly(A) tail in mRNA translation. Each category of proteins points to a location/region of poly(A)+ RNA where they most likely associate with when identified by RPL. PABPs, poly(A) binding proteins; CPA, cleavage and polyadenylation; TIFs, translation initiation factors; TEFs, translation elongation factors
