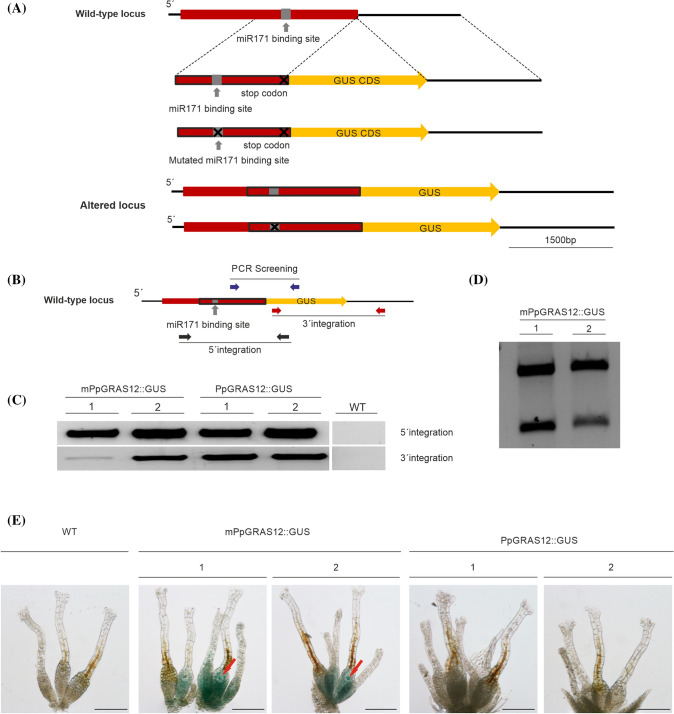
Fig. 1.
Generation of the PpGRAS12::GUS protein fusion reporter lines. a Scheme representing the generation of the PpGRAS12::GUS and mPpGRAS12::GUS fusion reporter constructs. Two variants of GUS fusion reporter constructs were generated and introduced to their cognate genomic locus by means of homologous recombination. The red box indicates the PpGRAS12 coding region. The red box with the black border lines indicates 1482 bp from the coding sequence including the miR171 binding site (native/mutated), which was fused to the GUS coding sequence (yellow box). The PpGRAS12 stop codon was removed and the coding sequence fused to the GUS coding sequence. b Purple, red, and black arrows show the primer pairs sequentially applied for PCR-based analyses of the PpGRAS12::GUS protein fusion reporter lines. c Upper panel: confirmation of 5′ integration of the constructs using black primers. Lower panel: confirmation of 3′ integration of the construct using red primers. d Validation of mPpGRAS12::GUS protein fusion reporter lines by digestion of RT-PCR products with PauI (a PauI restriction site was introduced into the mutated miR171 binding site). e Correspondent blue colors were detected only in the archegonia and egg cells of mPpGRAS12::GUS protein fusion reporter lines. Red arrows: egg cells. Scale bars: 1 mm