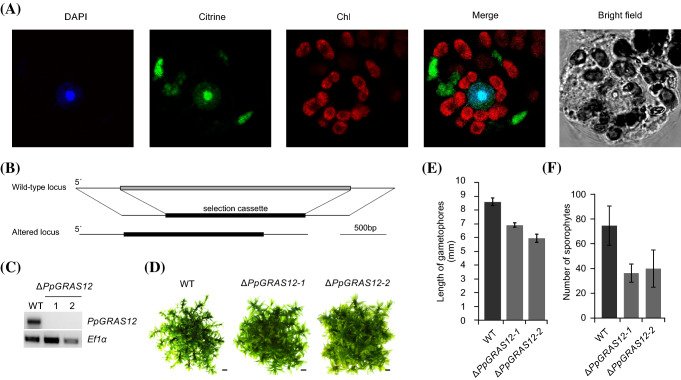
Fig. 2.
Generation and phenotypic analysis of the ΔPpGRAS12 lines. a Subcellular localization of the PpGRAS12::citrine protein fusion in P. patens protoplasts. Pictures were taken 3 days after transfection of the PpGRAS12::citrine fusion construct into P. patens protoplasts. DAPI: DAPI signal. Citrine: citrine signal. Chl: chlorophyll auto-fluorescence. Merge: merged images of citrine and chlorophyll auto-fluorescence. b Scheme depicting the targeted knockout approach of the PpGRAS12 coding sequence. c RT-PCR from cDNA derived from the indicated lines using PpGRAS12-specific primers; note that the two ΔPpGRAS12 mutant lines are null mutants lacking the PpGRAS12 transcript; RT-PCRs performed with primers for the constitutively expressed gene PpEf1α served as a control to monitor successful cDNA synthesis. d Phenotypic analyses of the knockout lines. Initially, a single gametophore from the indicated lines was cultured on standard growth medium and pictures were taken after 45 days of growth under standard growth conditions. Scale bars: 1 mm. e Comparison of the gametophore length in the WT and two independent ∆PpGRAS12 lines. Gametophore length was measured from colonies grown for 45 days under standard growth conditions; error bars represent standard errors (n = 30). f Comparison of the sporophyte numbers in the WT and two independent ∆PpGRAS12 lines; error bars represent standard errors (n = 27)