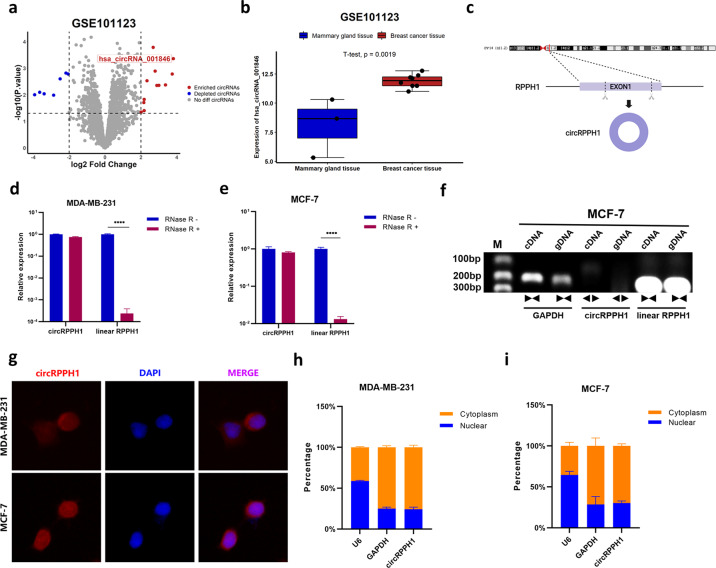
Fig. 1. The characteristics of circRPPH1 in BC.
a Bioinformatics analysis of GSE101123 identified differentially expressed circRNAs including hsa_circRNA_001846 (that is, circRPPH1). The cutoff was set at | log2(fold change) | > 2 and P < 0.05. b The expression of circRPPH1 was analyzed in BC tissue in GSE101123. c. CircRPPH1 was formed by backsplicing of RPPH1, exon 1. d–e The expression of circRPPH1 and linear RPPH1 were detected after treating total RNA of MDA-MB-231 (d) and MCF-7 (e) cells with RNase R. f The circular structure of circRPPH1 was identified by agarose gel electrophoresis. g The subcellular distribution of circRPPH1 was detected by FISH assay in MDA-MB-231 (up) and MCF-7 (down) cells. DAPI was used for nucleus staining. Blue, DAPI. Red, circRPPH1. h–i The levels of U6 (internal reference for nuclear transcripts), GAPDH (internal reference for cytoplasmic transcripts) and circRPPH1 were determined by RT-qPCR after subcellular fraction. *P < 0.05, **P < 0.01, *** P < 0.001, **** P < 0.0001.