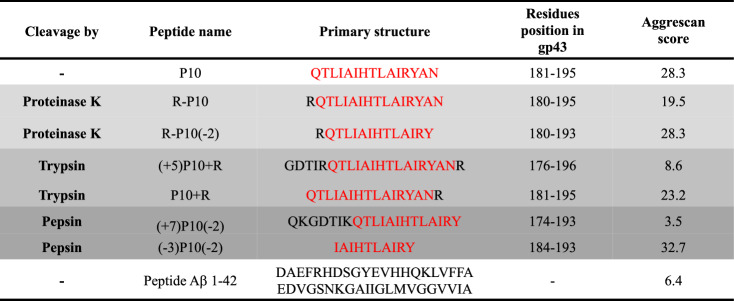
Table 1.
In-silico enzymatic digestion of gp43 shows that P10-like peptides can be formed.
Full gp43 primary sequence was subjected to the ExPASy PeptideCutter tool to identify possible cleavage sites. Proteinase K, trypsin and pepsin were the enzymes predicted to cleave gp43, generating P10-like peptides, while conserving P10 core sequence (HTLAIR). The peptide sequences and their positions in gp43 are depicted in the Table, where: R-P10 = P10 with an extra R at the N-terminal; R-P10(−2) = the same as R-P10 shortened two residues at the C-terminal; (+5)P10 + R = P10 with five extra residues at the N-terminal and R at the C-terminal; P10 + R = P10 with an extra R at the C-terminal; (+7)P10(−2) = P10 with seven residues at the N-terminal and shortened two residues at the C-terminal, and (−3)P10(−2) = P10 shortened three residues at the N-terminal and two residues at the C-terminal. In all sequences, P10 is colored in red. Aggrescan was used to estimate the aggregation propensities of the peptides. Peptide Aβ 1–42 was included for comparison.