Fig. 6.
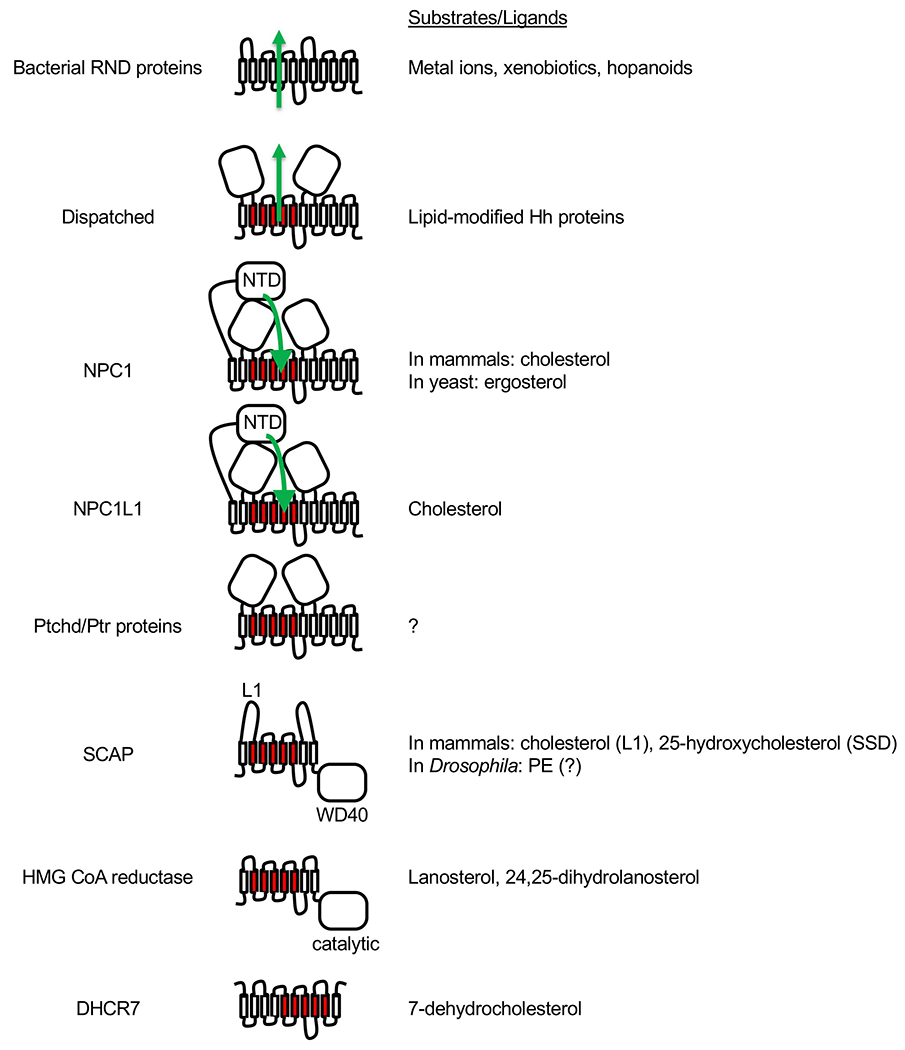
Bacterial RND proteins and eukaryotic SSD family members. For all proteins, the extracellular or luminal side faces up, and the cytosolic side faces down. The five TM domains that constitute the SSD are shown in red. Dispatched, NPC1, NPC1L1 and Ptchd/Ptr proteins are thought to function as transporters similar to the bacterial RND proteins, and the direction of substrate transport (where known) is indicated with green arrows. NPC1 and NPC1L1 each has an additional N-terminal domain (NTD). In the case of NPC1, cholesterol or ergosterol is first transferred from the luminal NPC2 protein (not shown) to the NTD and then travels through a tunnel between the other two large luminal domains towards the SSD. SCAP and HMG CoA reductase each has 8 TM domains and an additional cytosolic domain. In the case of SCAP, the cytosolic WD40 domain binds sterol regulatory element-binding proteins (SREBPs). Binding of either cholesterol to loop 1 (L1) or of 25-hydroxycholesterol to SSD of mammalian SCAP causes retention of SCAP-SREBP complex in the ER. Drosophila SREBP pathway responds to phosphatidylethanolamine (PE) instead of sterols but direct binding of PE to Drosophila SCAP has not been demonstrated. In the case of HMG CoA reductase, the cytosolic domain is necessary and sufficient for its catalytic activity, which is inhibited by statins. The membrane domain mediates ER-associated degradation of HMG CoA reductase in the presence of excess sterols. Lanosterol and 24,25-dihydrolanosterol have been shown to induce HMG CoA reductase degradation without affecting SREBP pathway. The topology of DHCR7 is not certain as it is predicted to contain 9 or 10 TMs, but its SSD is presumably the catalytic site where 7-dehydrocholesterol binds as a substrate.
