Figure 1: Nrxn1α mRNA is decreased in SD-Nrxn1tm1Sage HET pups and is not detectable in SD-Nrxn1tm1Sage KO pups.
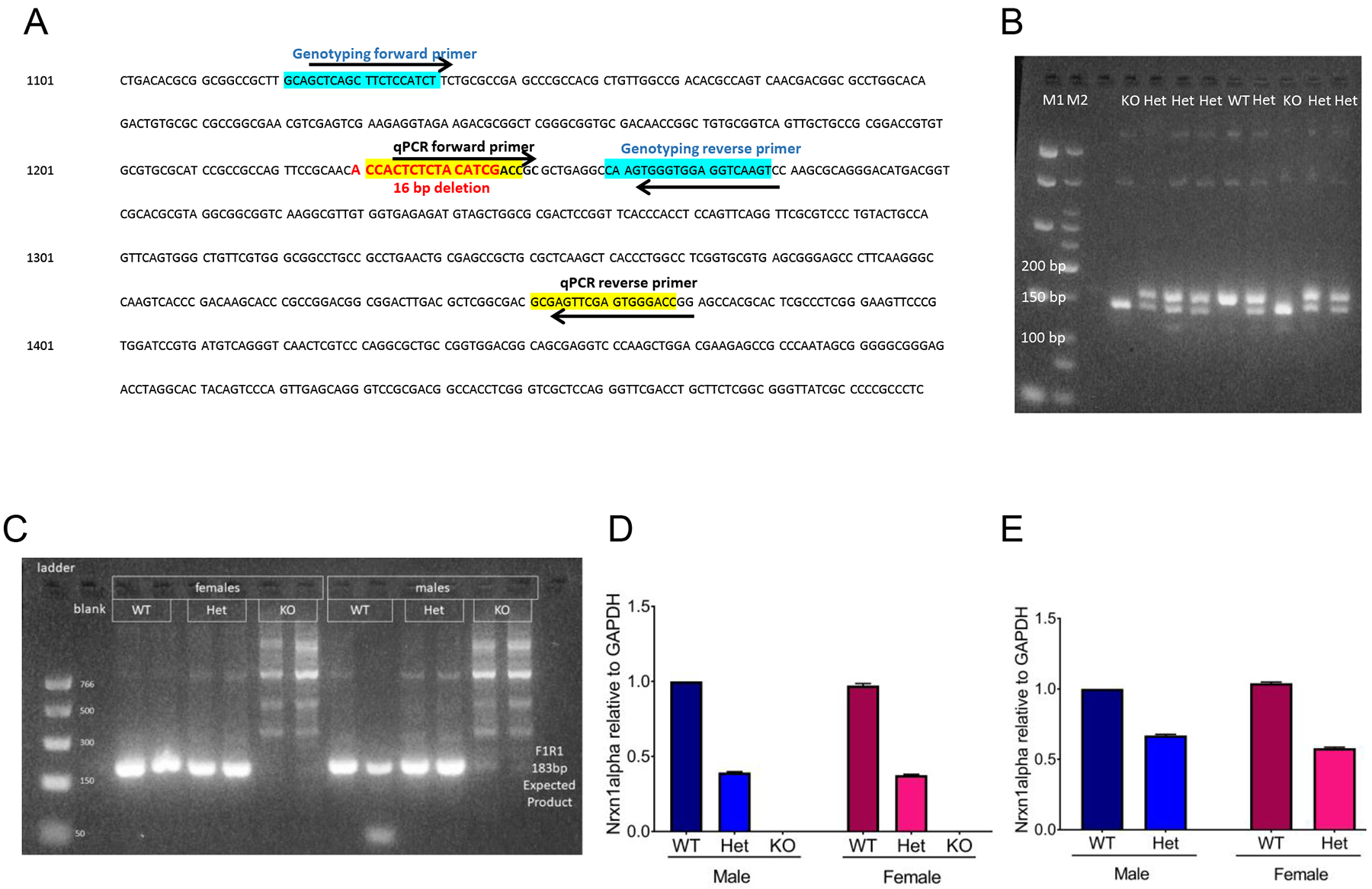
(A) Nrxn1α coding sequence. The region of exon 1 within the Nrxn1α gene that is deleted in the SD-Nrxn1tm1Sage rats is in bolded red text. The genotyping forward and reverse primers are highlighted in blue. The forward and reverse primers used for real-time quantitative PCR (qPCR) are highlighted in yellow. (B) Example agarose gel with products from a genotyping PCR reaction. (C–E) Nrxn1α expression in SD-Nrxn1tm1Sage pups containing wild type (WT), heterozygous (Het) and homozygous (KO) mutant Nrxn1α alleles as shown by qPCR. (C) Genomic DNA from tailsnips of rats of known genotype was used to validate qPCR primers for Nrxn1α expression. An 183bp product was expected for the WT Nrxn1α, which was detected in WT and Het males and females, but not in KOs of either sex. (D,E) Expression of Nrxn1α in cerebellum (D) and anterior cingulate cortex (E) of P2 pups, as determined by qPCR. Nrxn1α expression is normalized to Gapdh expression using the ΔΔCt method and expressed as mean fold expression relative to WT males +/− 95% confidence interval of mean fold expression. N = 6/group.
