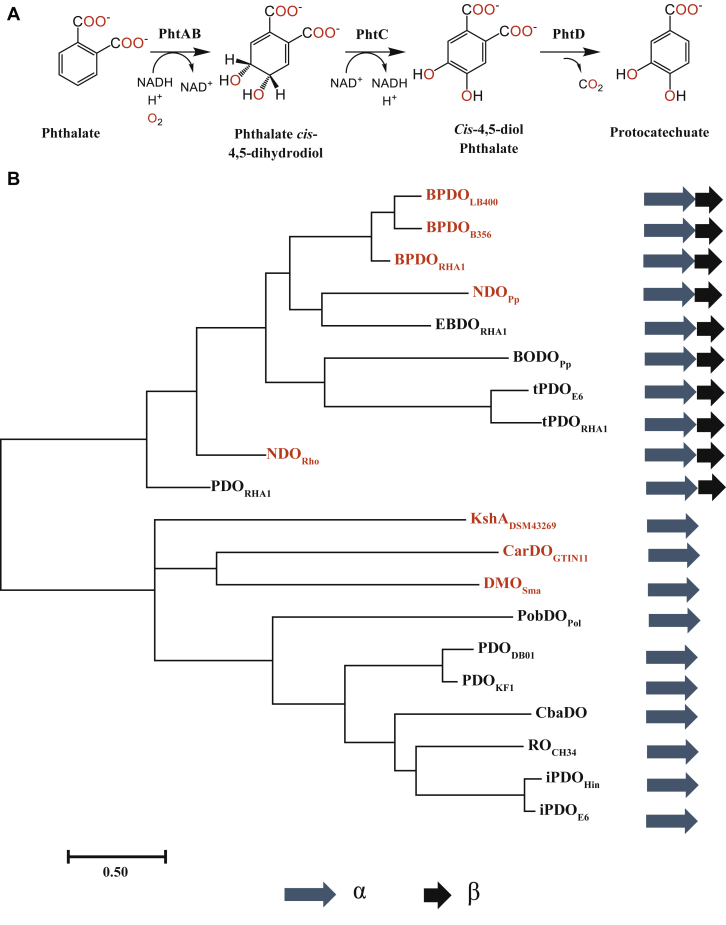
Figure 1.
Phthalate catabolic pathway and phylogeny of select ROs.A, catabolism of phthalate to protocatechuate by the Pht enzymes of C. testosteroni KF1. PhtA and PhtB correspond to PDO and PDR, which catalyze the 4,5-dihydroxylation of phthalate. Protocatechuate is catabolized to TCA cycle intermediates. B, phylogenetic relationship of PDOs. Unrooted phylogenetic tree was calculated using a sequence alignment of 20 Rieske oxygenase α-subunits. The proteins are abbreviated using the protein name and strain as follows (GenPeptID): biphenyl dioxygenase from Comamonas testosteroni B356 (BPDOB356, AAC44526), Paraburkholderia xenovorans LB400 (BPDOLB400, AAB63425), and Rhodococcus jostii RHA1 (BPDORHA1, BAA06868); ethylbenzene dioxygenase from R. jostii RHA1 (EBDORHA1, BAC92718); naphthalene dioxygenase from Pseudomonas putida 9816-4 (NDOPp, P0A110), and Rhodococcus sp. NCIMB 12038 (NDORho, AAD28100); terephthalate dioxygenase from Comamonas testosteroni E6 (tPDOE6, AIJ48578), and R. jostii RHA1 (tPDORHA1, ABH00392); benzoate dioxygenase from Pseudomonas putida (BODOPp, WP_011600771); carbazole dioxygenase from Sphingomonas sp. GTIN11 (CarDOGTIN11, AAL37976); 3-ketosteroid 9α-hydroxylase from Rhodococcus rhodochrous DSM43269 (KshADSM43269, B6V6V5); dicamba monooxygenase from Stenotrophomonas maltophilia (DMOSma, AAV53699); phthalate dioxygenase from Comamonas testosteroni KF1 (PDOKF1, EED67076), Rhodococcus jostii RHA1 (PDORHA1, BAD36800) and Burkholderia cepacia DB01 (PDODB01, WP_011881604); 3-chlorobenzoate dioxygenase from Comamonas testosteroni BR60 (CbaDO, Q44256); phenoxybenzoate dioxygenase from Pseudomonas oleovorans (PobDOPol, Q52185); RO from Cupriavidus metallidurans CH34 (ROCH34, ABF12407); isophthalate dioxygenase from Comamonas testosteroni E6 (iPDOE6, BAH70269), and Hydrogenophaga intermedia (iPDOHin, CDN90362). Structurally characterized proteins are marked with red.