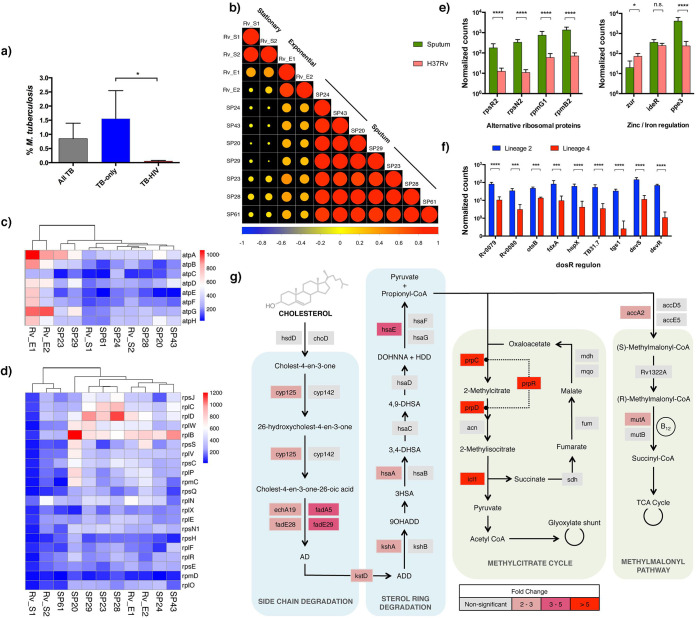
FIG 3.
Transcriptional profiles of sputum M. tuberculosis. (a) Despite active TB disease, M. tuberculosis only accounted for 0.85% ± 2% of all mapped bacterial reads. The percentage of M. tuberculosis reads was, however, significantly higher in TB-only samples than in TB-HIV samples (n = 9 and n = 8, respectively; P < 0.05 [Mann-Whitney U-test]). (b) The differential gene expression between seven sputum M. tuberculosis samples and laboratory cultures was calculated using DESeq2. The expression data were plotted as a correlation matrix with hierarchical clustering. Exponential cultures were labeled as Rv_E1 and Rv_E2, stationary cultures were labeled as Rv_S1 and Rv_S2, and sputum samples started with the initials SP. A decrease in circle size indicates reduced correlation; red indicates a positive correlation, and blue indicates a negative correlation. The sputum samples showed a high degree of concordance to each other and correlated more closely to exponential-phase cultures than to stationary-phase cultures. (c) Transcript abundance of ATP synthase genes in sputum M. tuberculosis clusters more closely to stationary-phase H37Rv than to exponential-phase cultures. (d) In contrast, transcript abundance of the two major ribosomal protein operons S10 and L14 in sputum M. tuberculosis were found to be more similar to exponential-phase H37Rv than to stationary-phase cultures. The color of the heatmaps corresponds to the normalized read count of each gene. (e) Significantly higher expression of four zinc-independent alternative ribosomal proteins was detected, along with decreased expression of the zur repressor and upregulation of ppe3, indicating that sputum M. tuberculosis was zinc deprived. (f) Expression of selected members of the DosR regulon. Consistent with the presence of an alternative transcriptional start sites in lineage 2 isolates (22), the transcript abundance of the DosR genes was significantly higher in lineage 2 sputum than in lineage 4 sputum M. tuberculosis. (g) Compared to exponential-phase laboratory cultures (H37Rv), M. tuberculosis in sputum was found to have significantly higher expression of 34 members of the KstR and KstR2 regulons associated with cholesterol catabolism and 6 members of the downstream propionate detoxification pathways. A pathway map is shown here to illustrate the transcript expression of some of the enzymes involved in the processes. Genes that were not differentially expressed (nonsignificant) are colored in gray, and those that were differentially expressed in sputum were colored in a scale of pink and red colors according to their fold change. No downregulated genes were identified in the KstR/KstR2 regulons or either of the propionate detoxification pathways. For panels e to f, adjusted P values (P-adj) were determined by DESeq2 and are indicated by asterisks (*, P-adj < 0.05; **, P-adj < 0.01; ***, P-adj < 0.001; ****, P-adj < 0.000; n.s., not significant).