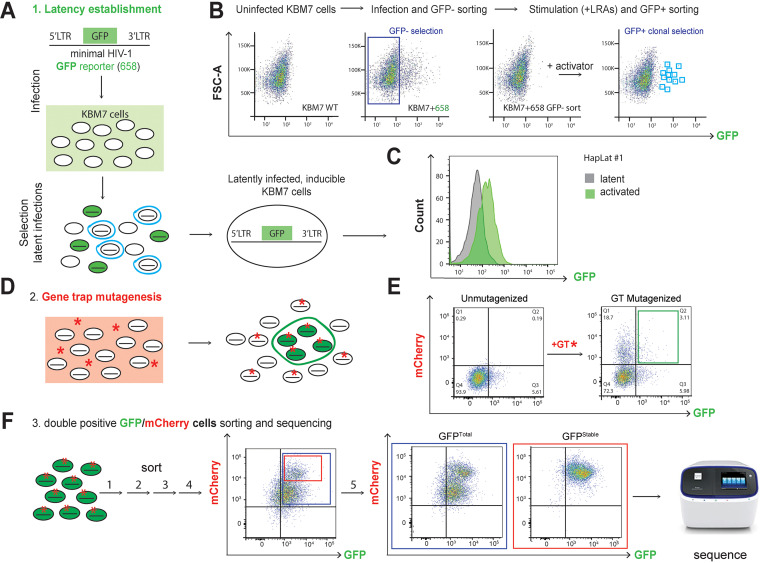
FIG 1.
Schematic representation of the two-color haploid screening strategy for identification of novel host factors and cellular pathways involved in the maintenance of HIV-1 latency. (A) Scheme depicting the generation of a clonal latent haploid KBM7 cell line. To this end, haploid KBM7 cells were infected with a minimal HIV-1 virus harboring GFP (HIV-1 658). After stimulation, reactivated cells were sorted and left to expand and revert to a latent state. (B) FACS plots depicting the establishment of haploid latent HIV-1 infected KBM7 cell line (Hap-Lat). Parental KBM7 cells were infected with a minimal HIV-1 virus carrying a GFP reporter (HIV-1 658). GFP-negative cells, consisting of uninfected and latently infected cells, were sorted by FACS. The polyclonal cell pool was stimulated with a cocktail of latency reversal agents (LRAs), and reactivated cells were clonally sorted by FACS and expanded to generate haploid latent cell lines. WT, wild type. (C) Hap-Lat #1 displays low basal activity (GFP expression) but is effectively reactivated using LRAs. (D) Hap-Lat cells were mutagenized by infection with a gene-trap (GT) virus harboring an mCherry reporter. Cells infected with the GT reporter will be mCherry positive (red asterisk). Latently infected KBM7 cells that reactivate following GT mutagenesis will be double positive for GFP and mCherry (green cells, red asterisk). (E) Representative FACS plots demonstrating gating strategy for sorting double-positive cells (GFP, mCherry). (F) Double-positive cells (GFP, mCherry) are sorted in multiple rounds to eliminate cells stochastically reverting to a GFP-negative state. During these rounds of sorting, a stable and distinct double-positive subpopulation (GFP Sub) appears which was sorted separately.