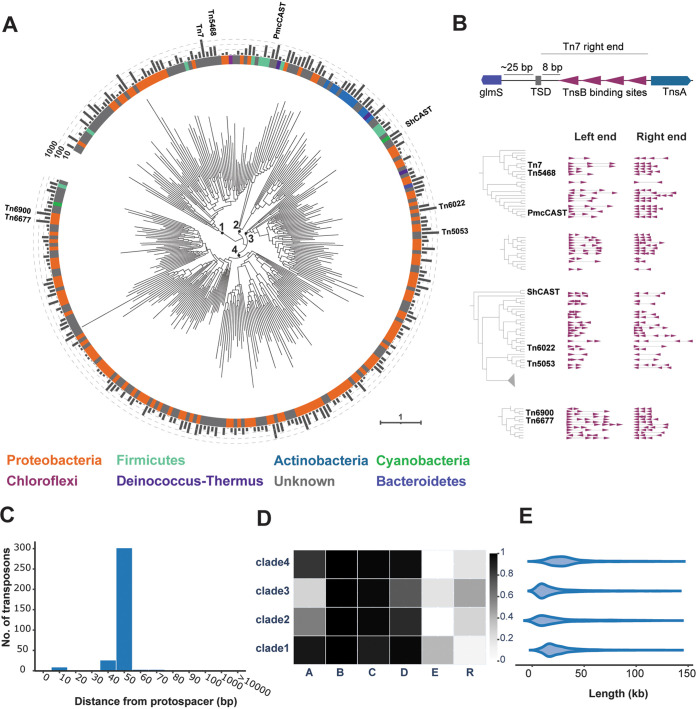
FIG 1.
Tn7-like transposons are widespread mobile genetic elements across the diversity of bacteria. (A) Phylogenetic tree of the DDE-family TnsB transposase from Tn7 and related transposons (n = 299 leaves). Deep branches are marked for clarity, and the number of transposases represented by each leaf is plotted on the outer ring. The taxonomic phylum for each leaf is plotted on the inner ring and assigned using the consensus of ≥80% of the transposase sequences; otherwise, the leaf phylum is marked “unknown.” (B) The predicted boundaries from selected transposons are displayed for a TnsB subtree, highlighting the fact that related transposases possess similar TnsB binding site architectures. Each binding site is depicted as an arrow, where sites downstream of the transposase were arbitrarily oriented to the positive strand (left end, arrows pointing right) and sites upstream of the transposase are on the negative strand (right end, arrows pointing left). (C) Histogram of distances from the predicted boundary of CASTs to the protospacer-targeted attachment site. (D) Heat map of the presence/absence of Tn7 core proteins. (E) Length distribution of intact, dereplicated transposons (n = 6,549).