Figure 4. In silico characterization of the SNARE complexes bearing the Asp68His and the Ile51Ser variant.
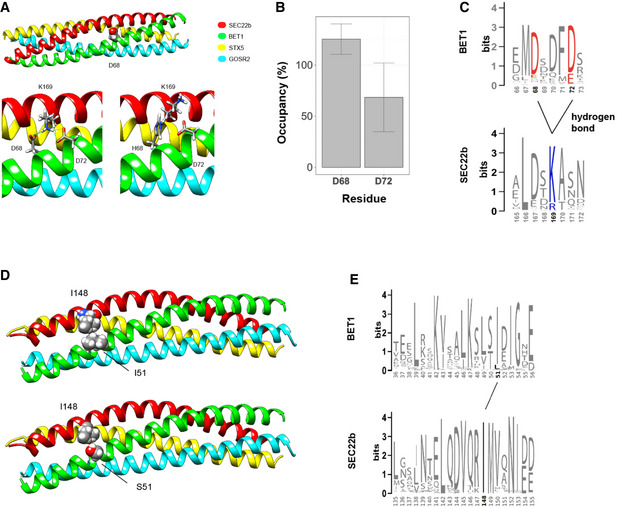
- Representation of the SNARE complex with Asp68 highlighted (top). BET1Asp68 is involved in a hydrogen bond with SEC22bLys169, with the most probable conformation of His68 in the mutant variant (bottom).
- Stability of hydrogen bonds between BET1Asp68 and BET1Asp72 respectively was estimated using molecular dynamics simulation in three replicas. Parameter occupancy could be more than 100% if several stable hydrogen bonds could be established by residue. Bars represent average fraction of the simulated time the residue is involved in a hydrogen bond(s). Data represented are mean ± SD.
- Sequence conservation of the corresponding regions of BET1 and SEC22b animal proteins. The relevant residues involved in hydrogen bond network are highlighted by color, hydrogen bonds are shown.
- Representation of the SNARE complex with Ile51 (upper) and Ser51 (lower) residues highlighted.
- Sequence conservation of the corresponding regions of BET1 and SEC22b and the relevant residues involved in the hydrophobic patch formed by Ile51 BET and SEC22bIle148.
