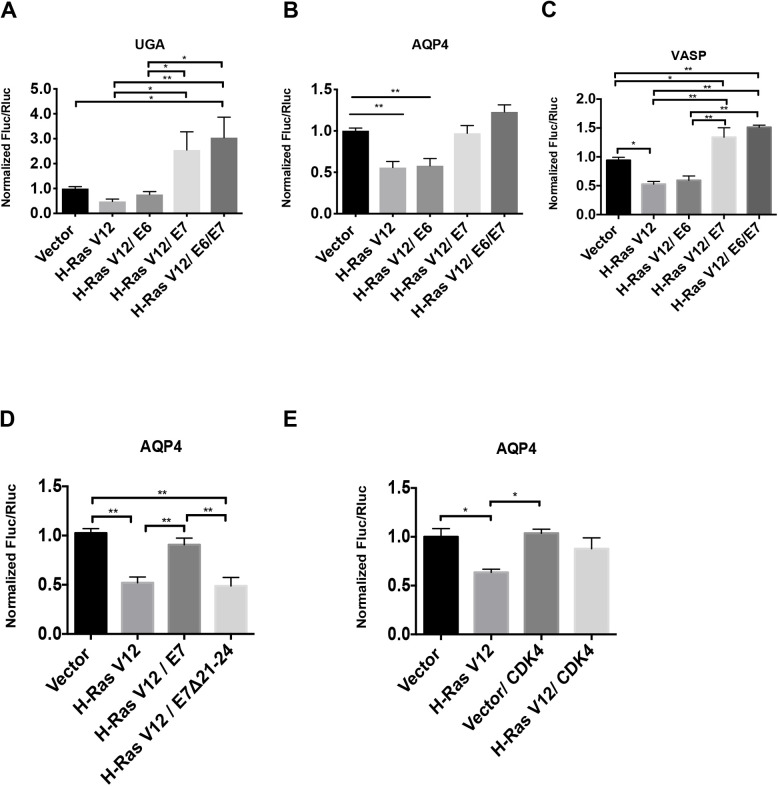
Fig. 4.
RB pathway disruption increases readthrough. (A–C) Readthrough efficiencies expressed as normalized Fluc/Rluc obtained from the luciferase reporters bearing a UGA stop codon within an artificial context (A) or a stop codon within natural contexts from either AQP4 (B) or VASP (B) that were expressed in IMR-90 cells. These cells also expressed HTERT, an empty vector (Vector) or H-RasV12 and an empty control vector (pLXSN), E6, E7 or E6/E7 oncogenes to study the readthrough efficiency in proliferating versus senescent versus transformed-like cells, n=3. (D) Relative readthrough luciferase values from IMR-90 cells transduced with the AQP4 readthrough reporter, with an empty vector (Vector) or H-RasV12 and an empty control vector (pLXSN), wild-type E7 or E7 Δ21-24 mutant oncogene. (E) IMR-90 cells were transduced with an empty vector or H-RasV12 or CDK4 and AQP4 luciferase reporters to study the readthrough efficiency variations in proliferating versus senescent versus cells overexpressing CDK4, n=3. A–E. Luciferase activities were measured in non-senescent and senescent cells at day 12 post-infection. Normalizations are presented as means relative to vector-infected cells, n=3 with technical triplicates for each experiment. One-way ANOVA with post-hoc Tukey HSD were performed. Error bars indicate SD of biological triplicates. Tukey HSD P-values indicate that *=P<0.05, **=P<0.01 are significantly different.