Figure 1. Extraembryonic DNA: Hypomethylation and Gestational Age Regulation.
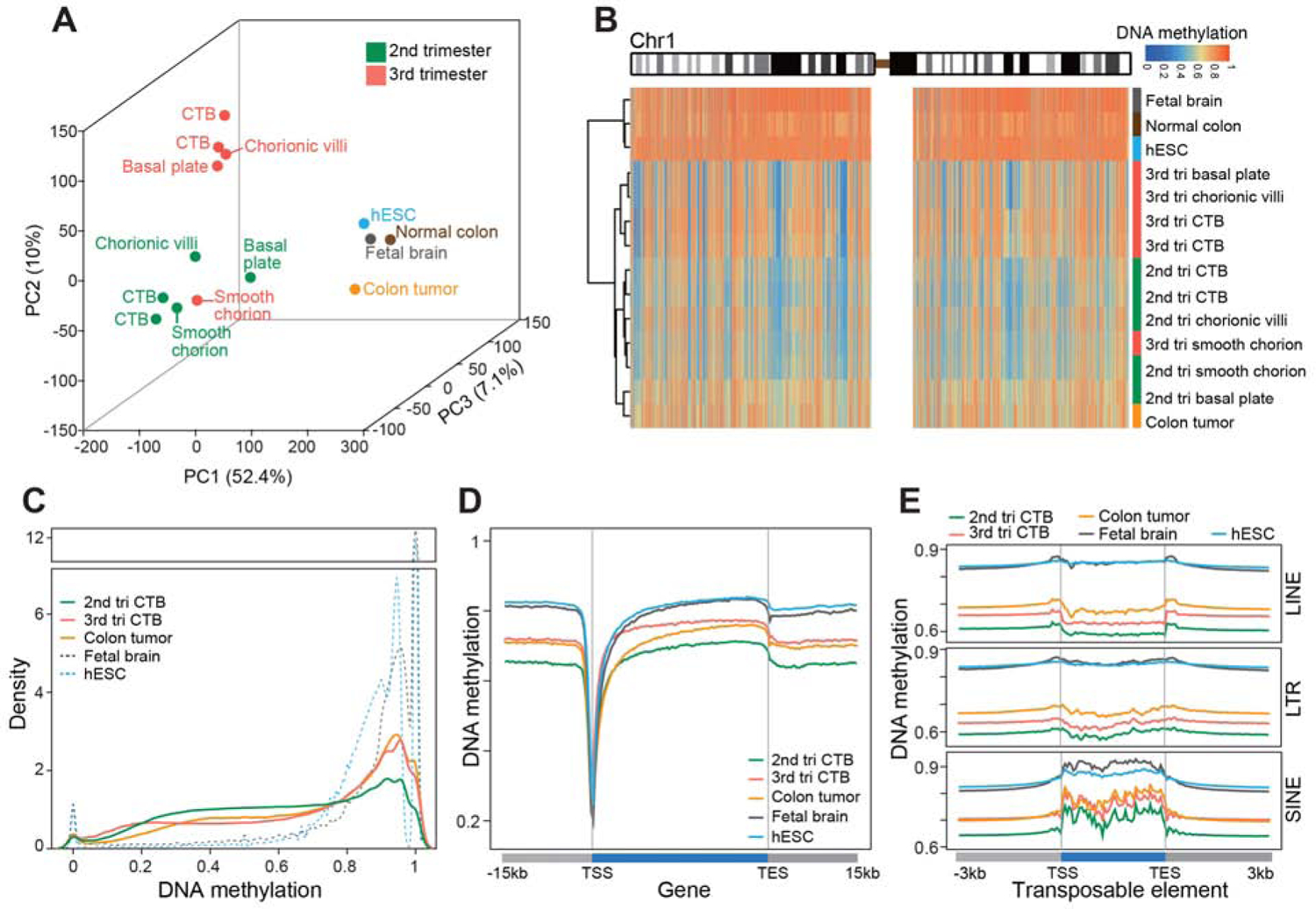
Whole genome bisulfite sequencing enabled quantitative single base-resolution DNA methylation profiling of the extraembryonic compartment. The data were compared to equivalent analyses of embryonic, fetal, and adult (normal and cancer) samples. (A) Principal component analysis segregated the extraembryonic samples from all others primarily along PC1. Within the extraembryonic group, the samples separated according to type and gestational age. The exception was the smooth chorion, which contains a cytotrophoblast progenitor population pregnancy. (B) Chromosome-level (Chr1) view of DNA methylation in the same samples as shown in (A). Compared to the other genomes, extraembryonic DNA showed a unique pattern of global hypomethylation interspersed among megabase domains of deeper hypomethylation. Similarly, colon tumor DNA was hypomethylated, but in a different pattern. (C) As compared to the other embryonic and fetal samples, 2nd trimester CTBs had an intermediate level of DNA methylation. At term, the cells acquired higher levels of methylation similar to the colon tumor. (D–E) Averaged DNA methylation levels over +/− 15 kb regions of gene bodies (RefSeq) and +/− 3 kb regions of transposable elements showed the same trends as in (C). 2nd/3rd trimester CTBs, n=2; 2nd/3rd chorionic villi, smooth chorion, and basal plate, n=1; tri., trimester; TSS, transcription/transposon start site; TES, transcription/transposon end site.
