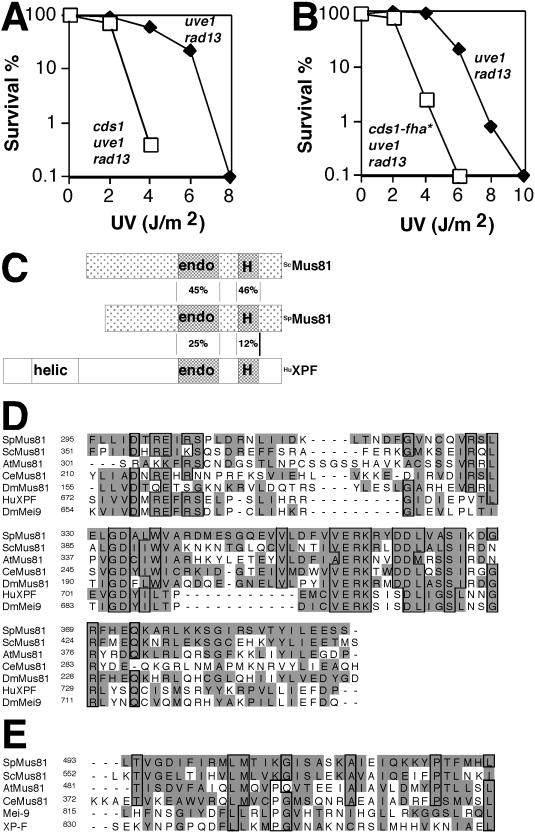
FIG. 1.
Identification of Mus81. (A) Cds1 is important for DNA damage tolerance. The uve1 rad13 and cds1 uve1 rad13 strains were assayed for UV survival (all results shown for UV sensitivity assays are representative examples of two or more experiments). (B) The cds1-fha* allele impairs DNA damage tolerance. uve1 rad13 and cds1-fha* uve1 rad13 cells were assayed for UV survival. (C) Schematic representation of Mus81 from S. pombe (SpMus81) and S. cerevisiae (ScMus81) and human XPF (HuXPF) (572, 632, and 905 amino acids, respectively). Percent identities between putative endonuclease (endo) and helix-hairpin-helix (H) domains are shown. The nonconserved helicase domain of XPF is shown (helic). Light shading depicts regions sharing more than 27% identity. (D) Alignment of endonuclease domains of Mus81 homologs and XPF family members. Sp, S. pombe; Sc, S. cerevisiae; At, Arabidopsis thaliana; Ce, Caenorhabditis elegans; Dm, Drosophila melanogaster, Hu, human. Shading highlights homologous residues, and the boxes show identities (PSI-BLAST results; alignment with ClustalW). (E) Alignment of the helix-hairpin-helix domains of Mus81 homologs and XPF family members.