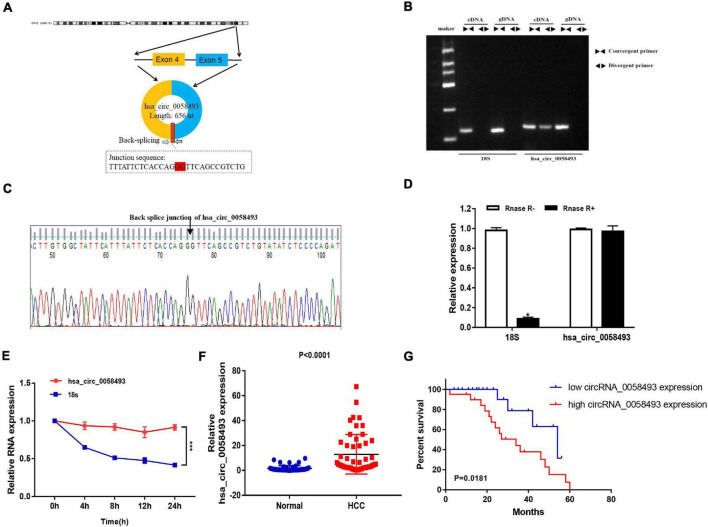
FIGURE 1.
(A) The circular structure of hsa_circ_0058493 showed that the hsa_circ_0058493 primer was derived from exons 4–5 of the RHBDD1 gene. Yellow represents exon4, blue represents exon5, and red represents the Back-splicing. (B) Gel electrophoresis results showed that the head-to-head primers can amplify products in both gDNA and cDNA, while the back-to-back primers can only amplify hsa_circ_0058493 in cDNA, which proved that hsa_circ_0058493 had a circular structure. (C) The reverse splicing junction of hsa_circ_0058493 was detected by Sanger sequencing. (D) The results of the RNase R enzyme digestion test showed that the expression of linear RNA decreased visibly after RNase R enzyme digestion, while the expression of hsa_circ_0058493 after RNase R enzyme treatment did not change clearly. (E) After treatment with actinomycin D in HCCLM3 cells, the relative RNA expression of hsa_circ_0058493 and mRNA were detected at different times. (F) Relative expression of hsa_circ_0058493 in 51 pairs of HCC and normal tissues. (G) The overall survival curve with low and high expression of hsa_circ_0058493 in HCC was analyzed by Kaplan–Meier. *P < 0.05, ***P < 0.001 vs. control group.