Figure 2. Co-crystal structure of tRNALys3 in complex with HIV-1 MA.
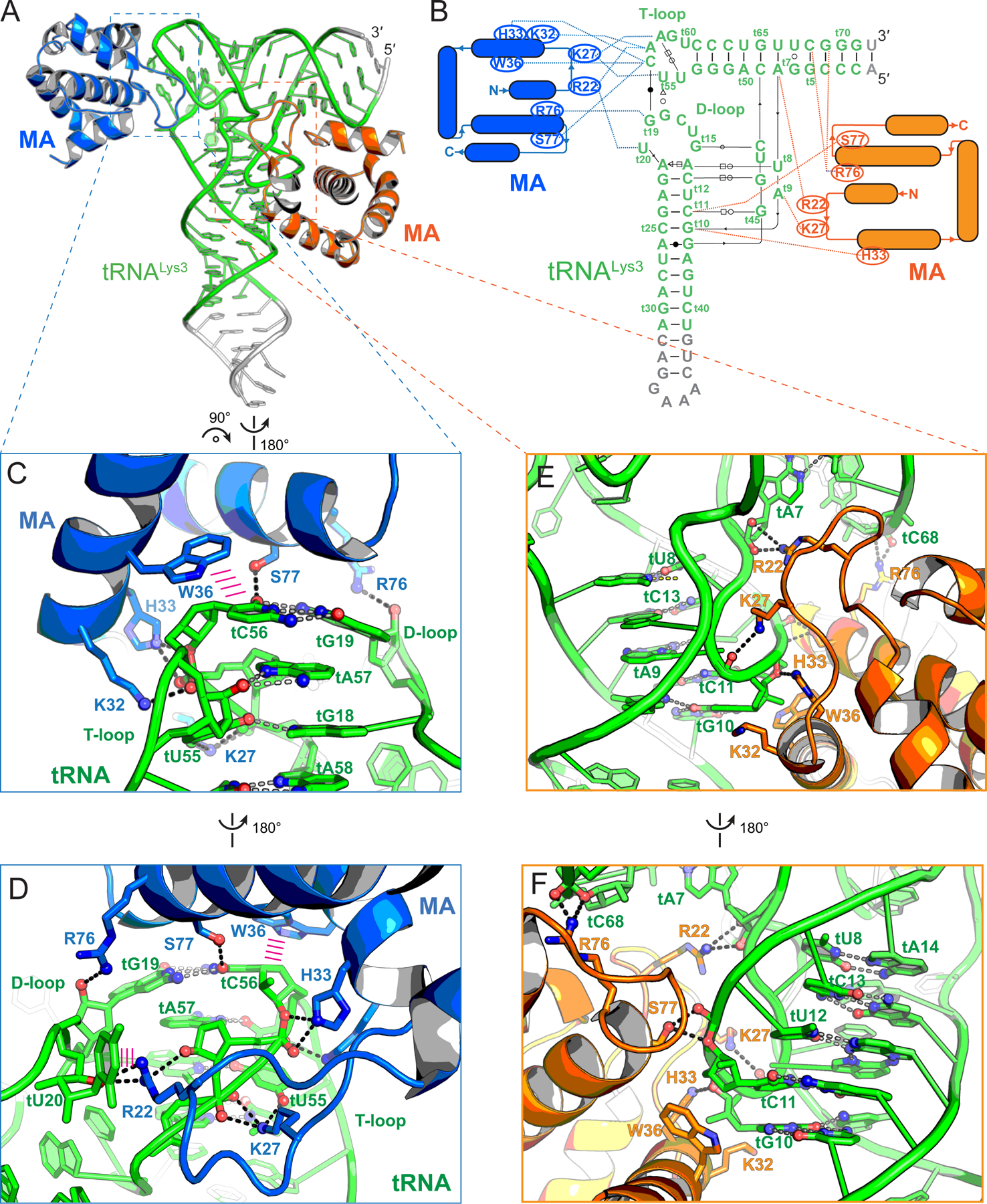
(A) Overall structure of tRNALys3 (green) bound to two MA molecules (blue and orange), which bind the tRNA elbow and inner corner regions, respectively. Engineered tRNA regions to promote crystallization shown in gray.
(B) Secondary structures and interactions of the MA-tRNALys3 complex. Noncanonical base pairs are indicated using Leontis-Westhof symbols (Leontis and Westhof, 2001). The tRNALys3 is numbered according to standard convention with a ‘t’ prefix. MA-tRNALys3 contacts as dashed lines.
(C & D) Specific recognition of tRNALys3 elbow structure by MA. Hydrogen bonds as dashed lines; stacking interactions as parallel magenta lines.
(E & F) Fortuitous interactions of tRNALys3 inner corner region with MA in crystallo.
