FIGURE 5. Locating the DWnt-5/Drl binding interface.
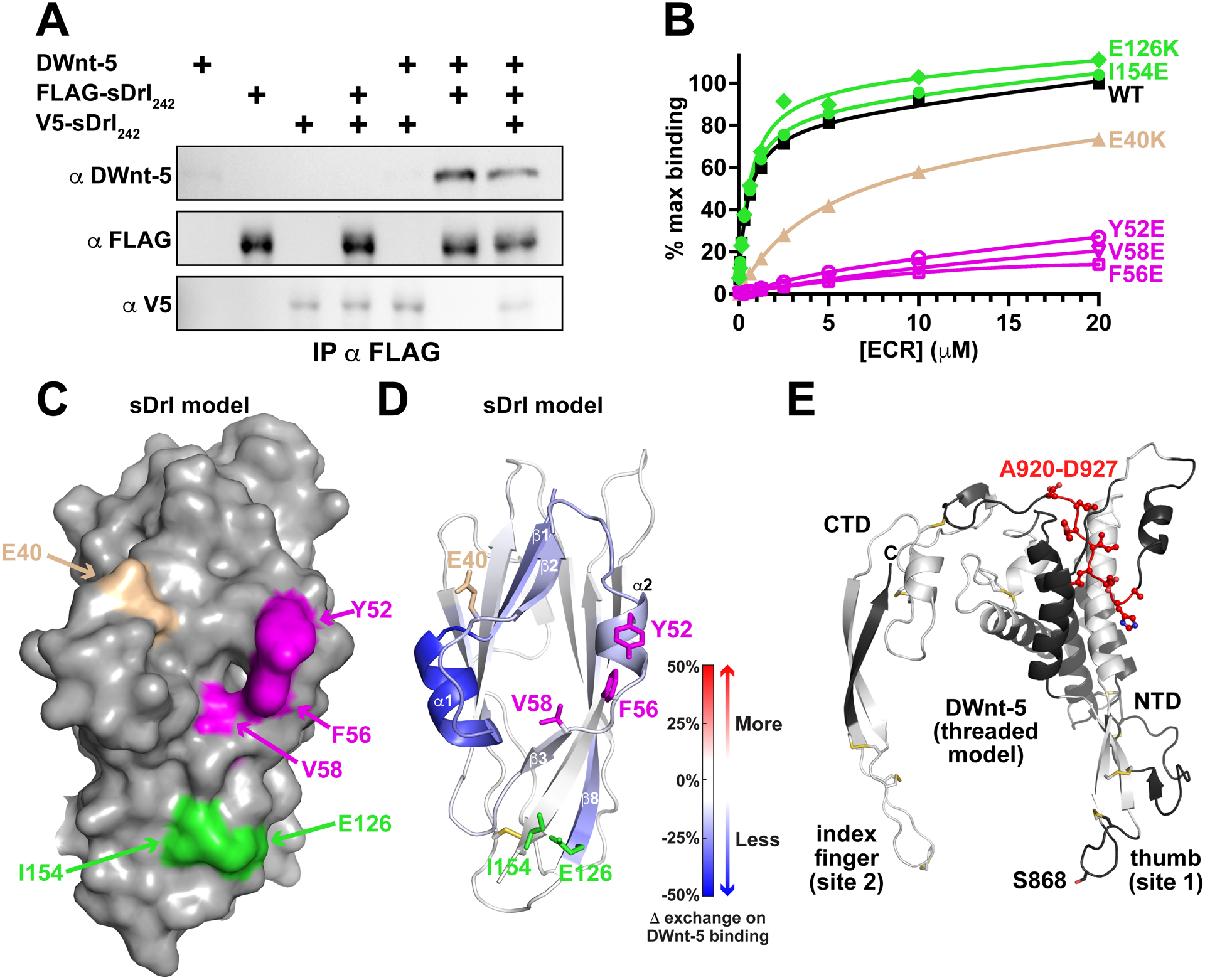
(A) Pull-down experiments indicate that DWnt-5 does not cause sDrl242 dimerization. FLAG-tagged and V5-tagged versions of sDrl242 were incubated with DWnt-5 (see Method Details) and immunoprecipitated with anti-FLAG. The quantity of V5-sDrl immunoprecipitated was not increased by the presence of FLAG-sDrl with- or without DWnt-5, arguing that they do not form dimers on WNT binding. Representative result from at least 3 independent experiments.
(B) SPR data showing that Y52E, F56E, and V58E mutations (magenta) in sDrl242 greatly impair binding to immobilized DWnt-5. An E40K mutation (tan) has an intermediate effect, whereas E126K or I154E mutations (green) have no detectable effect. Representative binding curves from at least 2 biological repeats are shown.
(C) Surface representation of the sDrl WIF domain model (based on the sDrl-2 WIF domain structure described here), showing the location of the mutations studied in B.
(D) Cartoon of sDrl model, colored by the change in ‘weighted relative difference’ in hydrogen/deuterium exchange (HDX) at the 1,000 s time point upon DWnt-5 binding (see Method Details). Regions in β1, α1, β2, α2 and to some extent β8 show some protection. Side-chains of residues mutated in B are marked and colored as in B.
(E) Limited HDX study of DWnt-5 changes upon binding sDrl in the same experiment. As described in the text, the large number of disulfides limit peptide coverage in DWnt-5. Grey/white areas are not seen in the recovered peptides. Only regions colored black were seen among the peptides (<50%), but none showed significant changes in HDX except the region colored red: A920-D927 in this threaded (xWnt8-based) model (Kelley et al., 2015) of DWnt-5.
See also Figure S5.
