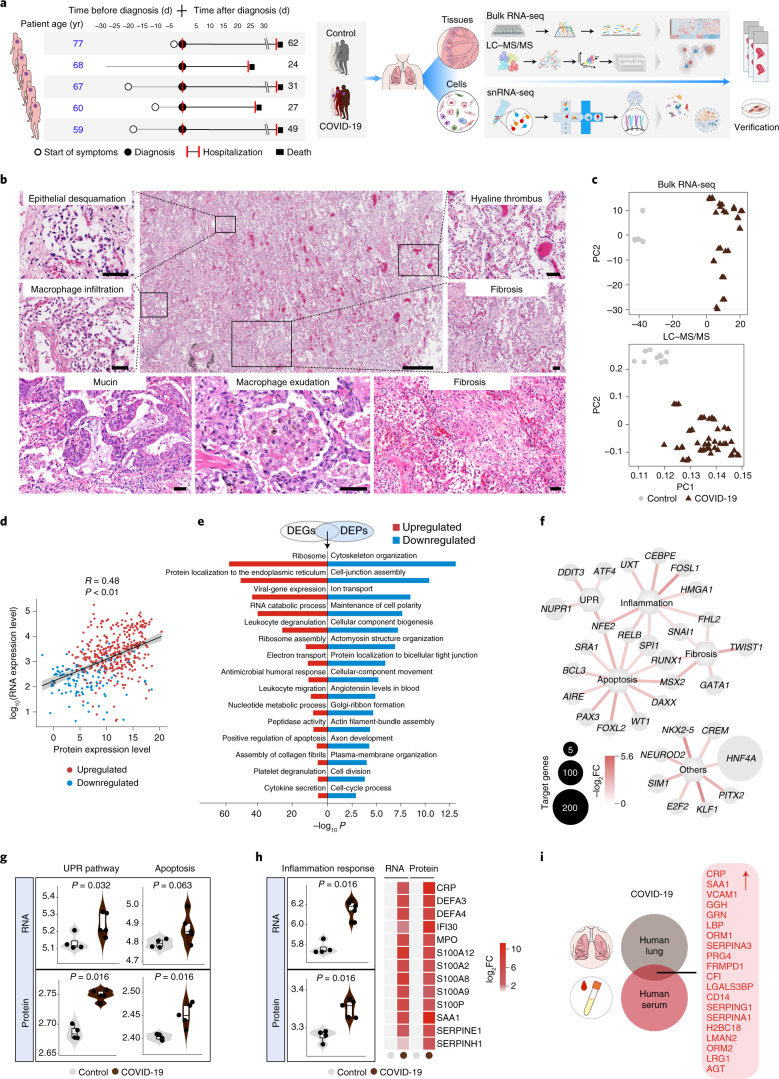
Fig. 1. Integrated analysis of COVID-19 pulmonary pathology.
a, Study overview. Information on the patients with COVID-19 and the pathological processes (left). Schematic of the experimental process of the analysis of five COVID-19 and four normal (Control) lung tissues by bulk RNA-seq, liquid chromatography with tandem mass spectrometry (LC-MS/MS), snRNA-seq and experimental verification (right). b, Representative images of haematoxylin-and-eosin staining of lung sections from patients with COVID-19 (n = 5 lungs with samples from three lung lobes each). Scale bars, 400 μm (main image; centre top) and 50 μm (magnified images). c, Principle component (PC) analysis showing the differences between the lungs of Controls and patients with COVID-19 based on the expression patterns from the RNA-seq (top) and LC-MS/MS analyses (bottom). d, Spearman’s correlation analysis of the expression levels of overlapping genes and proteins that were differentially expressed between the COVID-19 and Control lungs (|log2(fold change, FC)| > 1.5, adjusted P < 0.05). Linear fitting is indicated by a black line with confidence intervals represented in grey shading. e, GO term and pathway enrichment analysis of overlaps between COVID-19 DEGs and DEPs. f, Network plot showing upregulated transcription factors identified by ingenuity pathway analysis of the bulk RNA-seq. The small and large node sizes indicate low and high numbers of target genes, respectively. g, Gene-set scores of UPR pathway- and apoptosis-related genes at the RNA (top) and protein (bottom) level. h, Gene-set scores of inflammation response-related genes at RNA and protein levels (left). Heatmap showing inflammation-related genes upregulated at both the RNA and protein level (right). g,h, Control, n = 4 lungs; and COVID-19, n = 5 lungs, with samples from three lung lobes each. The boxes show the median (centre line) and the quartile range (25-75%), and the whiskers extend from the quartile to the minimum and maximum values. P values are indicated; Wilcoxon signed-rank test. i, Venn plot showing the DEPs that are common to the lungs and sera of patients with COVID-19. The upregulated proteins are listed (right).