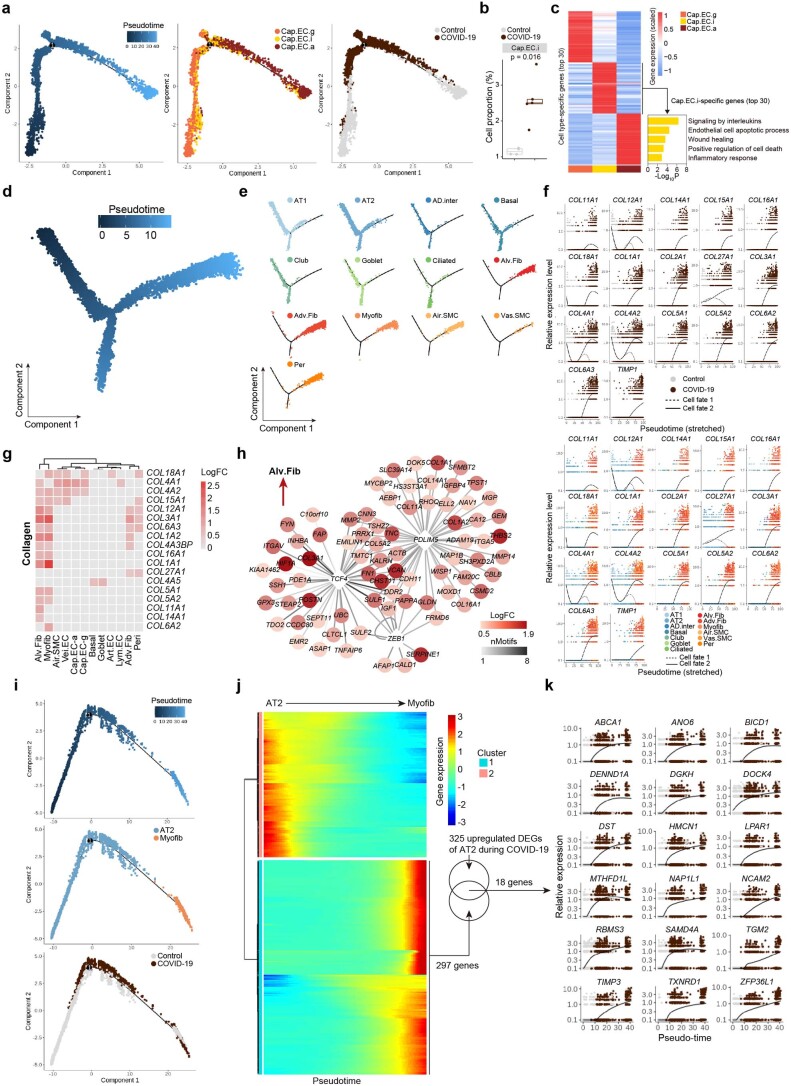
Extended Data Fig. 7. Transcriptomic signatures of endothelial cells and molecular hints for lung fibrosis in COVID-19.
(a) Left, pseudotime scores of capillary endothelial cells in human lung tissues. Middle, pseudotime trajectory of different capillary endothelial cell subtypes. Right, pseudotime trajectory of capillary endothelial cells from Control and COVID-19 samples. Cells are coloured by different cell types (middle) and groups (right). (b) Boxplot showing cell proportions of Cap.EC.i from Control (n = 4 donors) and COVID-19 (n = 5 donors) lungs. Box shows the median (centre line) and the quartile range (25-75%) and the whiskers extend from the quartiles to the minimum and maximum values. P values by Wilcoxon test are indicated. (c) Left, heatmap showing the gene expression signatures of the top 30 marker genes corresponding to each capillary endothelial cell type in human lungs. Right, bar plot showing the GO-term enrichment analysis of marker genes for Cap.EC.i. (d) Pseudotime scores of epithelial cells and stromal cells in human lung tissues. (e) Distributions of different epithelial cells and stromal cells in human lung tissues in pseudotime trajectory. (f) The relative gene expression patterns of the indicated genes over pseudotime of cell fate 1 (AT1) and cell fate 2 (myofib) in Control and COVID-19 lungs. Cells are coloured by groups (top) and different cell types (bottom). (g) Heatmap showing the expression signatures of collagen-related DEGs between COVID-19 and Control groups in different cell types. (h) Transcriptional network showing the core TFs identified in snRNA-seq from myofibroblasts by SCENIC analysis. (i) Pseudotime trajectory analysis of AT2 and myofibroblast in human lung coloured by pseudotime score, cell type and sample group. (j) Heatmap showing differentially expressed genes (q-value < 0.01) along the trajectory from AT2 to myofibroblast. (k) The relative gene expression patterns of the indicated genes along the pseudotime.