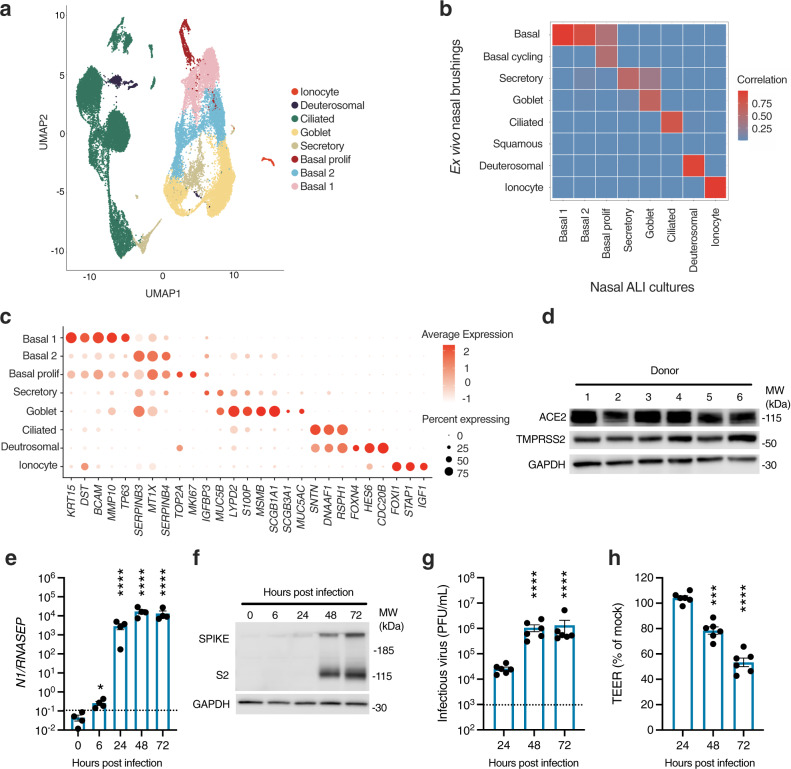
Fig. 1. Robust SARS-CoV-2 infection in a primary differentiated nasal epithelial ALI culture model.
a UMAP visualisation of single-cell RNA sequencing (scRNA-seq) data from nasal ALI cultures (28,346 single-cell transcriptomes from two representative donors) showed six major cell types. b Correlation between the annotation from an external dataset of nasopharyngeal swabs and the assigned annotation of our scRNA-seq from nasal ALI culture following label transfer. c Dot plot demonstrating expression of key markers distinguishing cell types in annotated clusters, with intensity demonstrated by colour and size of the dot representing the proportion of cells expressing the marker. d Immunoblot demonstrating ACE2 and TMPRSS2 expression by donor, representative of n = 3 experiments. Nasal ALI cultures were infected with SARS-CoV-2 (MOI 0.1) and subjected to various modalities to analyse infection. Whole-cell lysates were prepared at the indicated times for RT-PCR analysis of expression of e SARS-CoV-2 nucleocapsid (N1) gene expression normalised to the housekeeper RNASEP (average of n = 2 repeat experiments in n = 4 donors, mean ± SEM; *P = 0.0248, ****P < 0.0001, ANOVA, two-sided, with Dunnett’s post-test correction compared to 0 h). f Whole-cell lysates were prepared at the indicated times for immunoblot analysis of expression of SARS-CoV-2 spike (S) and cleaved S2 protein expression (representative of repeat experiments in n = 4 donors). g Release of infectious viral particles was measured by plaque assay of apical washings on permissive Vero E6 cells (average of repeat experiments in n = 6 donors, mean ± SEM; ****P < 0.0001, ANOVA, two-sided, with Dunnett’s post-test correction compared to 24 h). Dotted line represents lower limit of detection. h Transepithelial resistance (TEER) measurements upon infection (expressed as % of mock-infected wells, n = 6 donors, mean ± SEM; ***P = 0.0007, ****P < 0.0001, ANOVA, two-sided, with Dunnett’s post-test correction compared to 24 h). UMAP = Uniform Manifold Approximation and Projection, MOI = multiplicity of infection, PFU = plaque-forming units, MW = molecular weight, kDa = kilodalton, ACE2 = angiotensin-converting enzyme 2, TMPRSS2 = transmembrane serine protease 2, GAPDH = glyceraldehyde-3-phosphate dehydrogenase.