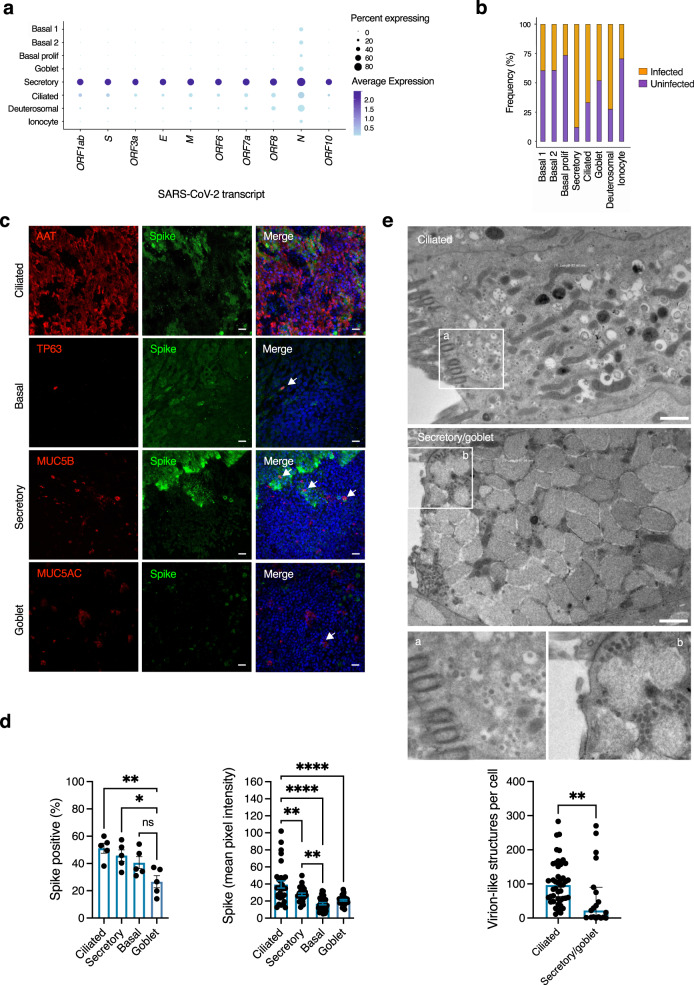
Fig. 2. Broad tropism of SARS-CoV-2 for nasal cells.
Nasal ALI cultures were infected with SARS-CoV-2 (MOI 0.1) and analysed using different modalities to explore tropism. At 24 h post-infection (hpi), cell suspensions were prepared from two representative donors for single-cell RNA sequencing (scRNA-seq) and 28,346 individual transcriptomes passing QC were analysed. a Dot plot of scRNA-seq data showing magnitude (colour) and proportion (size) of cell types expressing viral transcripts. E = envelope; M = matrix; N = nucleocapsid; S = spike, ORF = open reading frame. b Relative proportion of infected cell types based on expression of any viral transcript. Separately, nasal ALI cultures were fixed at 48 hpi and subjected to immunofluorescence analysis. c Expression of viral S protein expression in ciliated (AAT), basal cells (TP63), secretory (MUC5B) and goblet (MUC5AC) cells (arrowed) shown in (c). Scale bars = 10 μm (representative of experiments in n = 5 donors). d Quantification of cell-type specific expression of viral S protein (Goblet vs Secretory *p = 0.0309, Goblet vs Ciliated **p = 0.0045) and S protein intensity (Basal vs Secretory **p = 0.0073, Secretory vs Ciliated **p = 0.0046, ****p < 0.0001) at 48 hpi (n = 5 donors, mean ± SEM; ns = non-significant, ANOVA, two-sided, with Sidak’s post-test correction for multiple comparisons, indicated by lines). e Nasal ALI cultures were infected as above, fixed at 48 hpi for transmission electron micrograph (TEM) analysis of SARS-CoV-2 infected ciliated and secretory/goblet cells. Inserts a, b display virion-like structures in ciliated and secretory/goblet cells, respectively. Scale bars = 1 μm. Image analysis was undertaken to quantify virion-like structures as displayed in the bar plot (n = 3 donors, mean ± SEM **P = 0.0031, Mann–Whitney test, two-sided). MOI = multiplicity of infection, AAT = acetylated-alpha tubulin, tumour protein 63 = TP63, MUC = mucin.