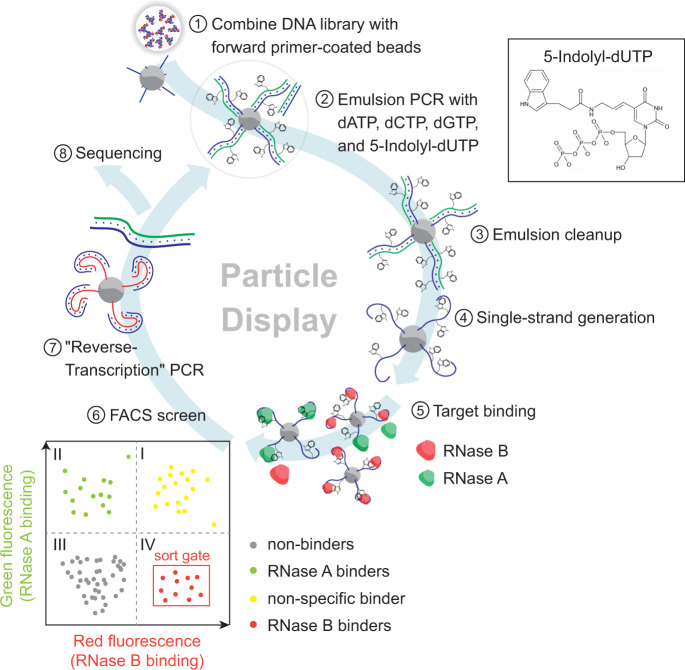
Fig. 1. A modified multiparameter particle display (MPPD) selection scheme for generating indole-modified aptamers.
1) DNA library molecules are hybridized to magnetic beads coated with forward primers, and 2) emulsion PCR is performed to create aptamer particles that each display many copies of a single sequence. dTTP is substituted with the modified base 5-indolyl-dUTP at this step to produce base-modified aptamers. 3) The emulsions are broken and 4) the aptamer particles are converted to single-stranded DNA via NaOH treatment. 5) The aptamer particles are incubated with RNase B (RB) and RNase A (RA), where each protein is labeled with a spectrally orthogonal fluorophore. 6) Fluorescence-activated cell sorting (FACS) separates aptamer particles that generate a strong RB-specific signal and minimal RA-specific signal (quadrant IV of the FACS plot). 7) The aptamers are subjected to a ‘reverse-transcription’ step to produce natural DNA, which is either used as the template for an additional round of screening or 8) characterized via high-throughput sequencing.