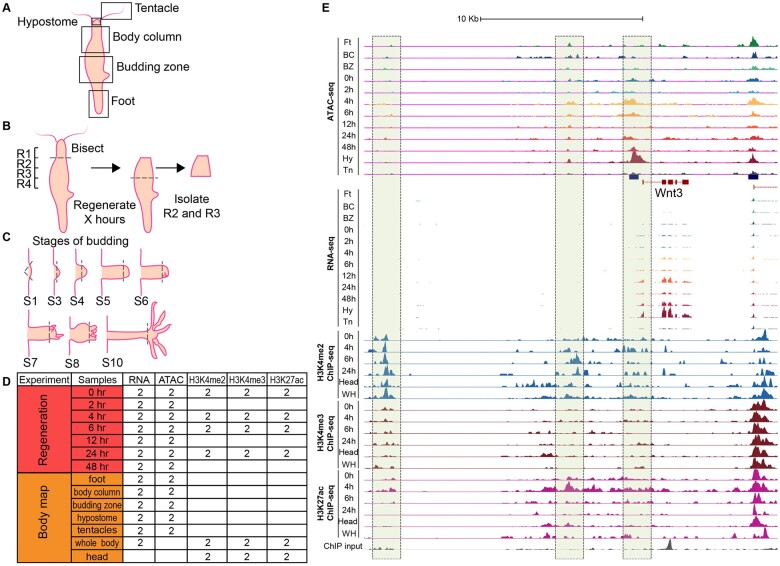
Fig. 1.
Outline of RNA-seq, ATAC-seq, and ChIP-seq experiments in Hydra. (A) Five Hydra body parts were isolated for RNA-seq and ATAC-seq to create a body map. (B) Hydra heads were bisected at the boundary of regions R1 and R2 and allowed to regenerate for specific time periods (0, 2, 4, 6, 12, 24, and 48 h). The regions R2 and R3 were isolated for RNA-seq to measure gene expression and ATAC-seq to map chromatin accessibility. (C) Hydra bud heads at various stages of budding (S1, S3, S4, S5, S6, S7, S8, and S10) were bisected and total RNA was extracted for RNA-seq. (D) Number of Hydra biological replicate samples assayed by RNA-seq, ATAC-seq, and ChIP-seq. Two biological replicates were also obtained for each stage of budding. (E) Genome browser signal tracks for RNA-seq, ATAC-seq, and ChIP-seq data for the Wnt3 locus. The Wnt3 promoter and an upstream “enhancer” region gain hypersensitivity along the head regeneration time course as gene expression is turned on.