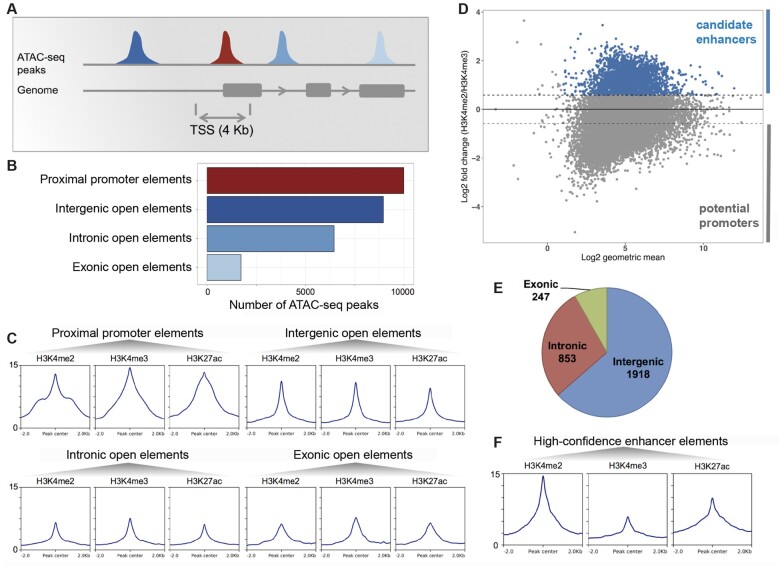
Fig. 4.
Identification of a high-confidence set of Hydra regulatory regions using ATAC-seq and ChIP-seq data. (A) Distribution of open-chromatin elements based on their genomic locations. (B) The 27,137 replicated ATAC-seq peaks found in one or more samples were classified based on overlap with gene loci. The proximal promoter regions were defined as regions within 2 kb of start of genes. (C) Enrichment of H3K4me2, H3K4me3, and H3K27ac histone modifications normalized signals at each type of open-chromatin elements. (D) About 3,018 open-chromatin elements (blue) with at least 50% higher H3K4me2 than H3K4me3 signal form a set of high-confidence candidate enhancer-like (in blue). Y axis shows log2-fold change of H3K4me2 signal over H3K4me3 signal and x axis shows the log2 of geometric mean of H3K4me2 and H3K4me3 signals. In the negative are potential promoters. (E) Genomic distribution of candidate enhancer-like elements. (F) Enrichment of H3K4me2, H3K4me3, and H3K27ac signals at the candidate enhancer-like elements.