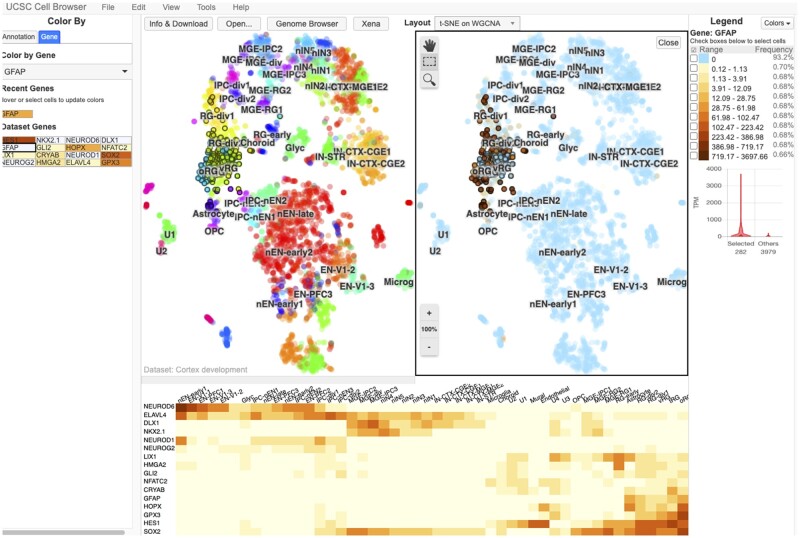
Fig. 1.
The UCSC Cell Browser interface showing a dataset focused on human cortex development. In the center of the screen, the primary layout view has been split into two vertical panes. The left pane shows the cells in this dataset colored by their metadata value for the field “WGCNACluster” (e.g. RG-div1). The right side shows the same cells colored by their expression of the gene HES1, those with a higher expression being colored dark red. On the far right side of the screen, there is a legend outlining how the colors are associated with expression bins; below this is a violin plot of this gene expression in a set of selected cells (those outlined in black in the center of the screen) versus all other cells in the dataset. On the far left side of the screen, you can see a list of metadata annotation fields available for this dataset. Each one can be clicked to color the plot by the values in that field. Below the two vertical panes, a heatmap shows 16 different genes that the authors of this dataset consider important “dataset genes”