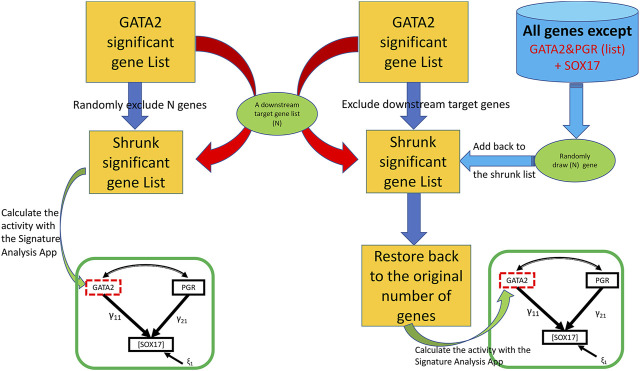
FIGURE 3.
A two-class bootstrap resampling (elimination with or without replacement) simulation. From the initial GATA2 significant gene list represented as the yellow rectangle, the downstream target genes (“N”) are eliminated in the without replacement simulation (left side) giving rise to the shrunk significant gene list represented by a smaller yellow rectangle; in the elimination with replacement simulation (right side), the same number of genes as that of the targeted subset of genes (“N”) are eliminated giving rise to the shrunk significant gene list, and then restored back to the original size by adding back randomly draw (“N”) represented by the far right green oval from the gene pool represented by the blue cylinder. In the elimination without replacement, the resulting shrunken GATA2 gene list is used to calculate the T-scores, then fed into the SEM model indicated by the green rectangle. In the elimination with replacement, the restored gene list is used to calculate the T-scores, then fed into the SEM model. The simulation can be repeated for a large “number of bootstraps” to generate a non-parametric distribution for statistics significance.