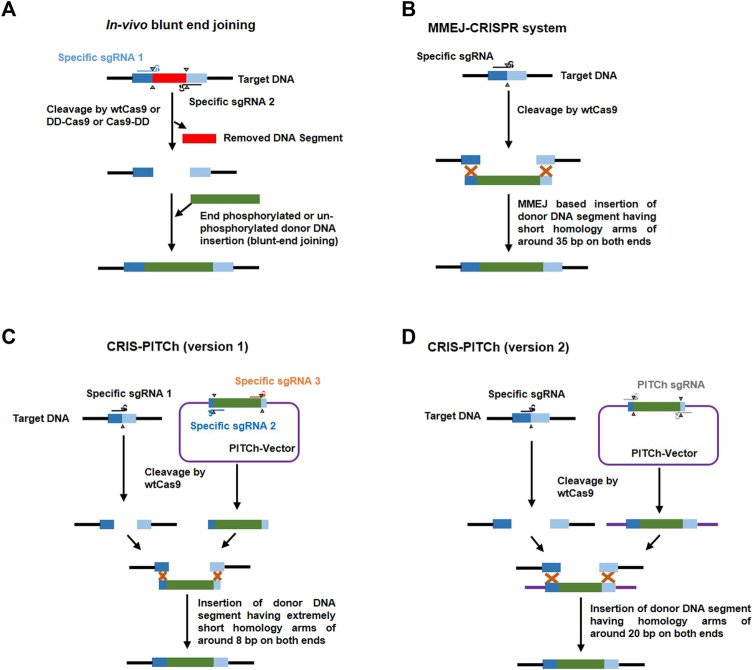
FIGURE 5.
Representative methods in donor-based host genome editing. (A) In vivo blunt-end joining involves using two target specific sgRNAs (blue and black) to cleave target DNA [releasing a DNA segment (red)] with either wildtype cas9 (wtCas9) or Cas9 attached to the protein destabilization domain (DD) like FKBP12-L106P which destabilizes Cas9 in the absence of a stabilizing agent, such as Shield-1 (Clontech); it leads to precise knock-in of donor DNA (green) via non-homologous end joining (NHEJ). (B) The MMEJ-CRISPR system involves cleavage of target DNA using one specific sgRNA (black) and wtCas9; and integration of a donor DNA fragment (with around 35 bp homologous arms at both ends (dark and light blue) at the cut site through the process of microhomology-mediated end joining (MMEJ) during the M-early S phases of the cell cycle. (C) CRIS-PITCh version 1 involves three specific sgRNAs [one to cleave target DNA (black) and the other two (yellow and brown) to cleave the DNA sequence flanking outside the homologous sequence (8 bp) in the donor vector (PITCh-vector) which releases a donor DNA segment with homologous arms (dark and light blue)] and wtCas9 to precisely integrate any gene of interest without end trimming by proximal MMEJ. (D) CRIS-PITCh version 2 also involves three sgRNAs [one specific sgRNA (black) for host target genome cleavage and two general PITCh sgRNAs (grey) to cleave the DNA sequence flanking outside the homologous sequence (20 bp) in the donor vector (PITCh-vector) which releases a donor DNA segment with homologous arms (dark blue and light blue)] and wtCas9 to precisely integrate any gene of interest with end trimming by distal MMEJ.