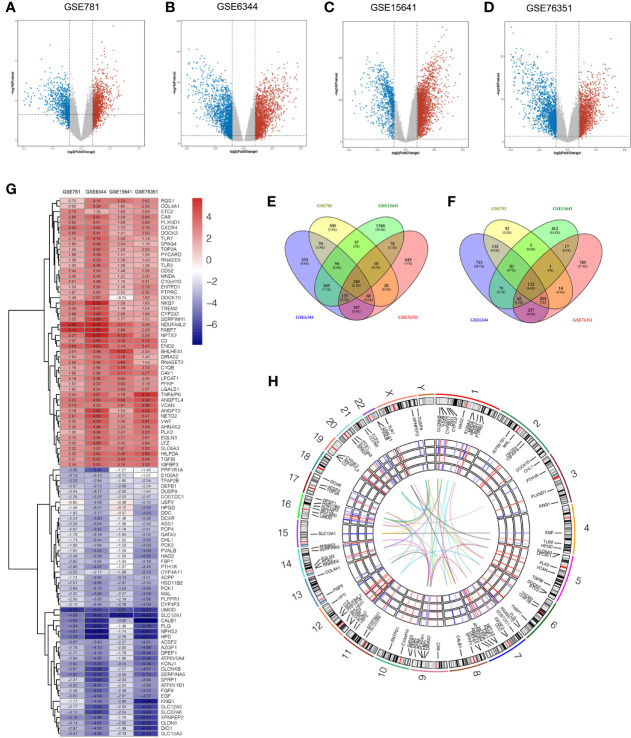
Figure 1.
Identification of DEGs between tumor and adjacent normal tissues in four ccRCC microarray datasets. (A–D) Volcano plots of GSE781 (A), GSE6344 (B), GSE15641 (C), and GSE76351 (D), from the GEO database. (E, F) Venn diagrams of the overlapping DEGs identified in the four datasets. The overlapping areas represent 240 upregulated genes (E), and 132 downregulated genes (F). The cutoff criteria were P value < 0.05 and |log2FC| > 1. (G) Heatmap of the top 50 upregulated genes and top 50 downregulated genes of DEGs. Each column represents a dataset and each row represents a gene. The number upon each rectangle is the log2FC value. (H) Circular visualization of expression profiles, chromosomal positions and correlation of the top 100 DEGs. The rainbow network in the center corresponds to a strong correlation (Pearson correlation coefficient > 0.8) among the top 100 DEGs. The four GEO datasets expression profiles are presented in the middle circular heatmaps. The gradient ranging from blue to red represents the changing spectrum from down- to upregulation. The outer circle corresponds to 24 chromosomes, and lines deriving from each gene point to their specific chromosomal locations. DEG, differentially expressed genes; ccRCC, clear cell renal cell carcinoma; GEO, Gene Expression Omnibus; log2FC, log2 Fold Change.