Figure 1.
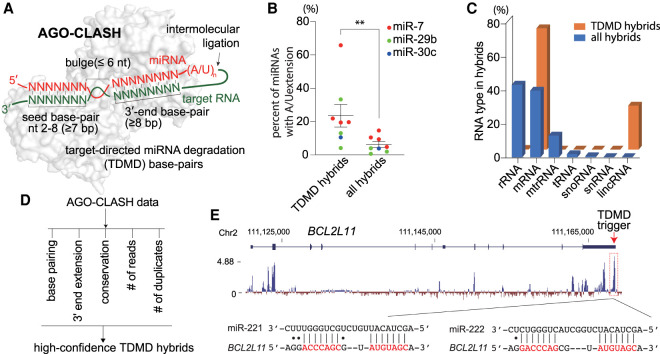
A wide range of transcripts contain potential TDMD triggers. (A) Schematic of AGO-CLASH and target-directed miRNA degradation (TDMD) base-pairing pattern. AGO2 structure (PDB: 6MDZ) is used in the illustration. A detailed description is given in the text. (B) The percentage of miRNA with extension in the hybrids of CYRANO/miR-7, NREP/miR-29b, SERPINE1/miR-30c, or all RNA/miRNA. Each dot represents a data point from one AGO-CLASH data set. (**) P < 0.01, ratio paired t test. (C) The composition of different types of RNA in the AGO-CLASH miRNA-containing hybrids obtained from HCT116 cells, comparing all hybrids and TDMD hybrids. (D) High-confidence TDMD hybrid screening based on five criteria. See the Materials and Methods for details. (E) Organization and conservation of the human BCL2L11 locus. Shown below the gene model ([blue boxes] exons, [blue line] introns) is a conservation plot, which is based on a 100-vertebrate basewise conservation by PhyloP. Diagramed below is the pairing of miR-221 and miR-222 to its potential TDMD trigger, located within the most conserved region of BCL2L11.