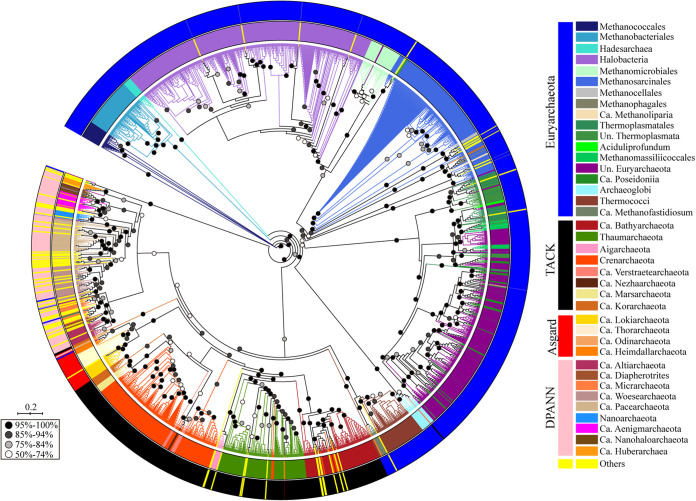
FIG 2.
Phylogenetic analysis of the aCPSF1 orthologs from 1,964 archaeal genomes/MAGs. In total, 1,964 aCPSF1 orthologs were retrieved from 2,520 available archaeal genomes/MAGs deposited in NCBI database. These genomes all have protein annotation information and phylogenetic affiliations. Un, unclassified; Ca, Candidatus; Others, unclassified and uncultured archaea. A maximum likelihood phylogenetic tree was constructed based on consensus amino acids of aCPSF1 protein sequences, inferred with FastTree v.2.1.10 under the WAG+GAMMA model (IQ-TREE 1.6.12 in the LG+C20+R4+F model, 1,000 ultrafast bootstraps replicates [52, 57, 58]), and visualized using iTOL v3 (59). Bootstrap supporting values of branch clustering are indicated by dots. Scale bar indicates number of substitutions per site.