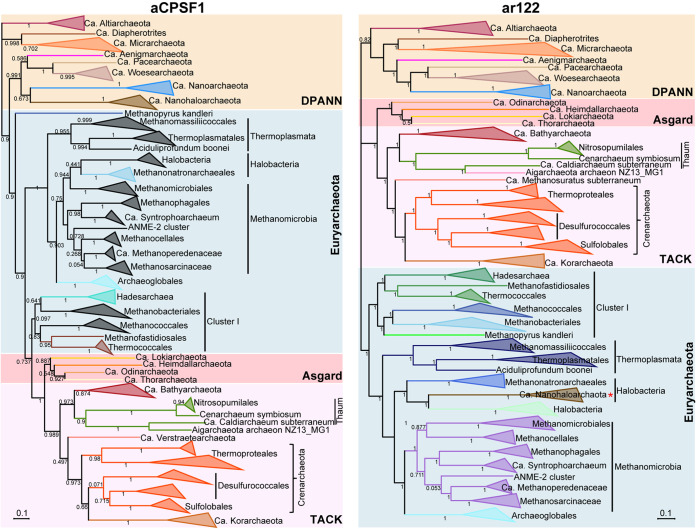
FIG 3.
Comparison of the aCPSF1 phylogenetic topology with the phylogenomic clustering of representative Archaea. The aCPSF1 orthologs and 122 archaeal marker proteins were retrieved from the same 143 representative cultured archaeal genomes or completed MAGs with high quality. Consensus sequences of the aCPSF1 proteins (left) and the concatenated 122 archaeal proteins (right) were used to construct the respective maximum likelihood phylogenetic trees using IQ-TREE (v.1.6.12) with “LG+I+G4” mode and 1,000 times ultrafast. The phylogenic trees were visualized with iTOL v3 (https://itol.embl.de/). Bootstrap evaluation values of 1,000 iterations are indicated at branch nodes. Scale bar indicates substitution numbers per site.