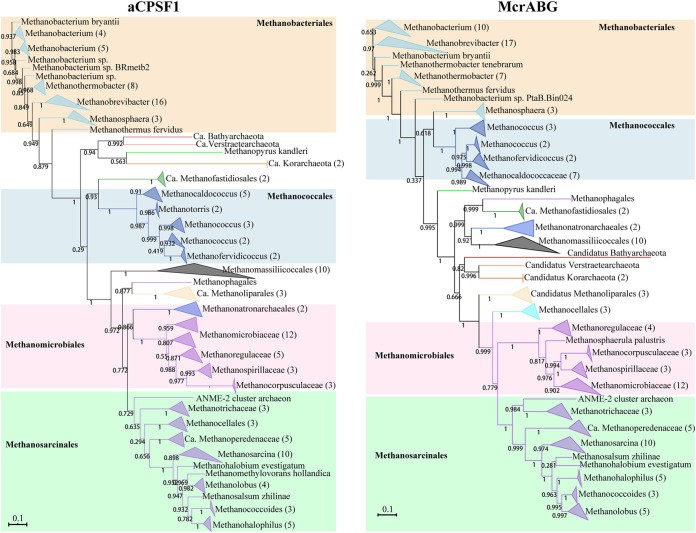
FIG 4.
Comparison of the phylogenetic trees of methanogenic archaea constructed based on aCPSF1 orthologs and the McrABG concatenation. In total, 138 methanoarchaeal genomes listed in Dataset S1 (“*” marked) representing all seven defined methanogenic orders and methanotrophic archaea were selected, and the aCPSF1 and McrABG orthologs were retrieved for phylogenetic study. The maximum likelihood phylogenetic trees were respectively constructed based on the protein sequences of aCPSF1 orthologs (left) and McrABG protein concatenation (right). Inside the parenthesis following the methanogens indicate the genome numbers from the same taxa. Bootstrap evaluation values of 1,000 iterations are indicated at branch nodes. Scale bar indicates number of substitutions per site.